Rushabh Patel
Graph Based Link Prediction between Human Phenotypes and Genes
Jun 01, 2021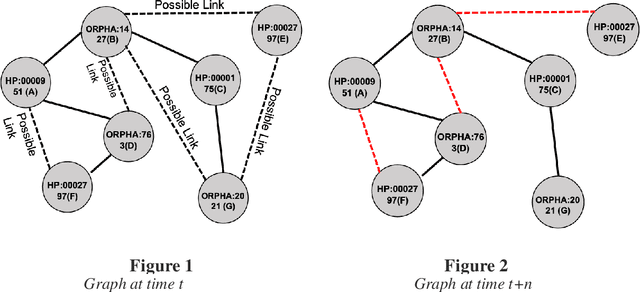
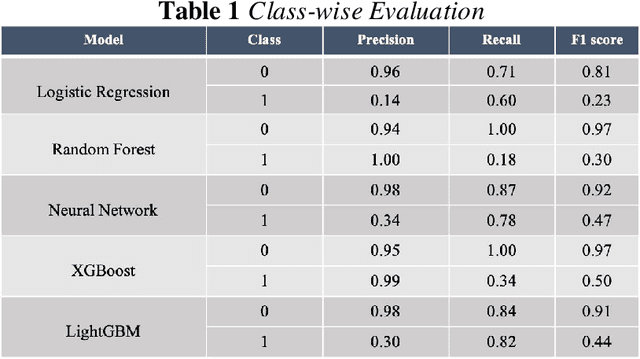
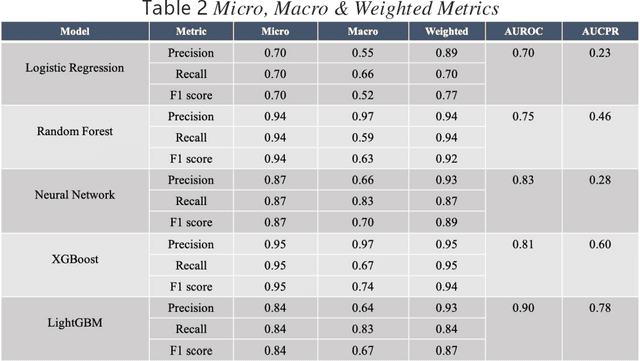
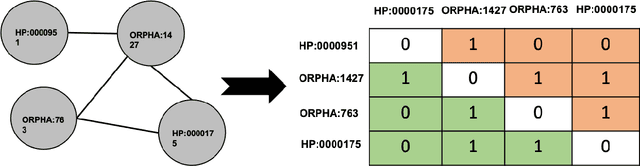
Abstract:Background: The learning of genotype-phenotype associations and history of human disease by doing detailed and precise analysis of phenotypic abnormalities can be defined as deep phenotyping. To understand and detect this interaction between phenotype and genotype is a fundamental step when translating precision medicine to clinical practice. The recent advances in the field of machine learning is efficient to predict these interactions between abnormal human phenotypes and genes. Methods: In this study, we developed a framework to predict links between human phenotype ontology (HPO) and genes. The annotation data from the heterogeneous knowledge resources i.e., orphanet, is used to parse human phenotype-gene associations. To generate the embeddings for the nodes (HPO & genes), an algorithm called node2vec was used. It performs node sampling on this graph based on random walks, then learns features over these sampled nodes to generate embeddings. These embeddings were used to perform the downstream task to predict the presence of the link between these nodes using 5 different supervised machine learning algorithms. Results: The downstream link prediction task shows that the Gradient Boosting Decision Tree based model (LightGBM) achieved an optimal AUROC 0.904 and AUCPR 0.784. In addition, LightGBM achieved an optimal weighted F1 score of 0.87. Compared to the other 4 methods LightGBM is able to find more accurate interaction/link between human phenotype & gene pairs.
Predicting invasive ductal carcinoma using a Reinforcement Sample Learning Strategy using Deep Learning
May 26, 2021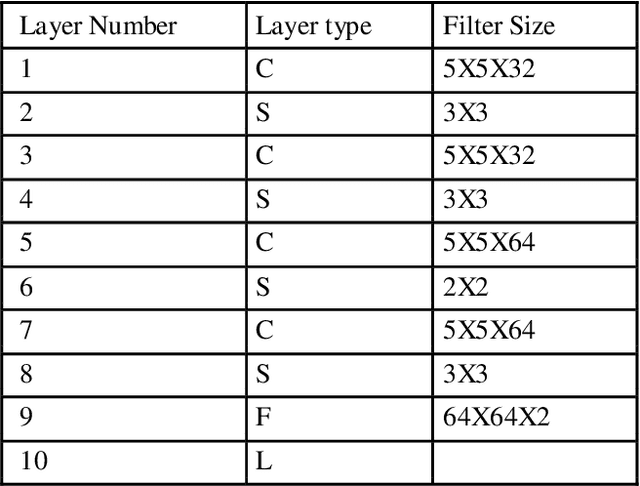
Abstract:Invasive ductal carcinoma is a prevalent, potentially deadly disease associated with a high rate of morbidity and mortality. Its malignancy is the second leading cause of death from cancer in women. The mammogram is an extremely useful resource for mass detection and invasive ductal carcinoma diagnosis. We are proposing a method for Invasive ductal carcinoma that will use convolutional neural networks (CNN) on mammograms to assist radiologists in diagnosing the disease. Due to the varying image clarity and structure of certain mammograms, it is difficult to observe major cancer characteristics such as microcalcification and mass, and it is often difficult to interpret and diagnose these attributes. The aim of this study is to establish a novel method for fully automated feature extraction and classification in invasive ductal carcinoma computer-aided diagnosis (CAD) systems. This article presents a tumor classification algorithm that makes novel use of convolutional neural networks on breast mammogram images to increase feature extraction and training speed. The algorithm makes two contributions.
Robotic Surveillance Based on the Meeting Time of Random Walks
Dec 04, 2019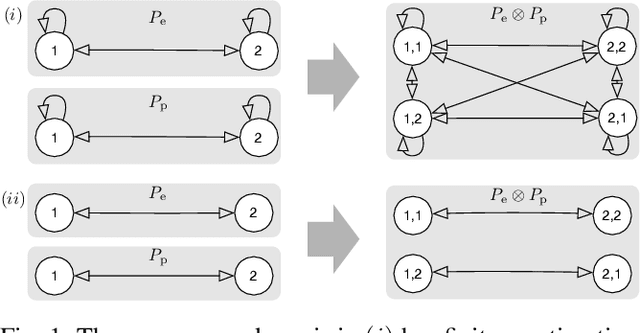
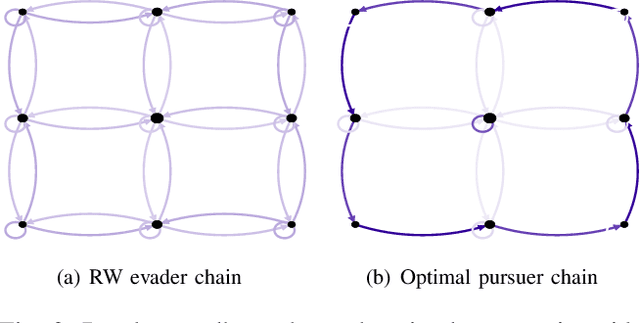
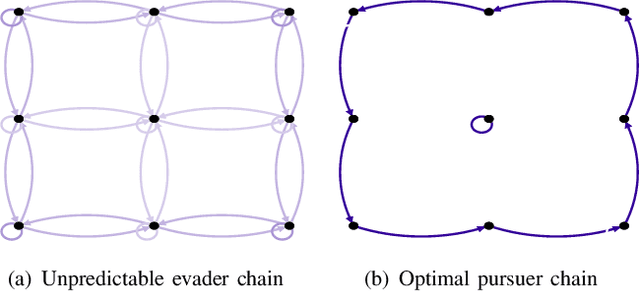
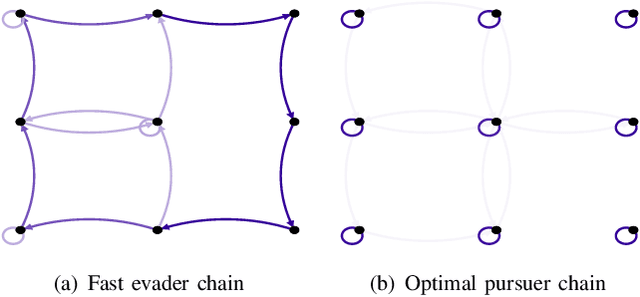
Abstract:This paper analyzes the meeting time between a pair of pursuer and evader performing random walks on digraphs. The existing bounds on the meeting time usually work only for certain classes of walks and cannot be used to formulate optimization problems and design robotic strategies. First, by analyzing multiple random walks on a common graph as a single random walk on the Kronecker product graph, we provide the first closed-form expression for the expected meeting time in terms of the transition matrices of the moving agents. This novel expression leads to necessary and sufficient conditions for the meeting time to be finite and to insightful graph-theoretic interpretations. Second, based on the closed-form expression, we setup and study the minimization problem for the expected capture time for a pursuer/evader pair. We report theoretical and numerical results on basic case studies to show the effectiveness of the design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge