Rosario Sarabia
Real-Time Brain Tumor Detection in Intraoperative Ultrasound Using YOLO11: From Model Training to Deployment in the Operating Room
Jan 27, 2025



Abstract:Intraoperative ultrasound (ioUS) is a valuable tool in brain tumor surgery due to its versatility, affordability, and seamless integration into the surgical workflow. However, its adoption remains limited, primarily because of the challenges associated with image interpretation and the steep learning curve required for effective use. This study aimed to enhance the interpretability of ioUS images by developing a real-time brain tumor detection system deployable in the operating room. We collected 2D ioUS images from the Brain Tumor Intraoperative Database (BraTioUS) and the public ReMIND dataset, annotated with expert-refined tumor labels. Using the YOLO11 architecture and its variants, we trained object detection models to identify brain tumors. The dataset included 1,732 images from 192 patients, divided into training, validation, and test sets. Data augmentation expanded the training set to 11,570 images. In the test dataset, YOLO11s achieved the best balance of precision and computational efficiency, with a mAP@50 of 0.95, mAP@50-95 of 0.65, and a processing speed of 34.16 frames per second. The proposed solution was prospectively validated in a cohort of 15 consecutively operated patients diagnosed with brain tumors. Neurosurgeons confirmed its seamless integration into the surgical workflow, with real-time predictions accurately delineating tumor regions. These findings highlight the potential of real-time object detection algorithms to enhance ioUS-guided brain tumor surgery, addressing key challenges in interpretation and providing a foundation for future development of computer vision-based tools for neuro-oncological surgery.
Postoperative glioblastoma segmentation: Development of a fully automated pipeline using deep convolutional neural networks and comparison with currently available models
Apr 17, 2024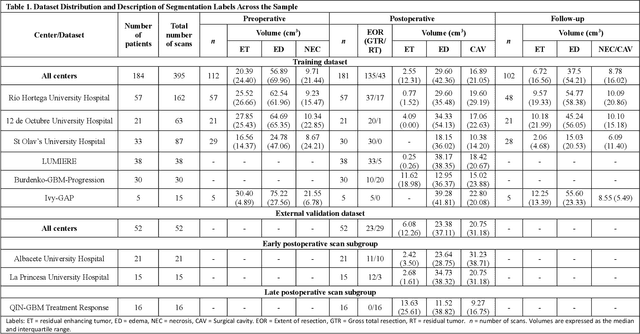
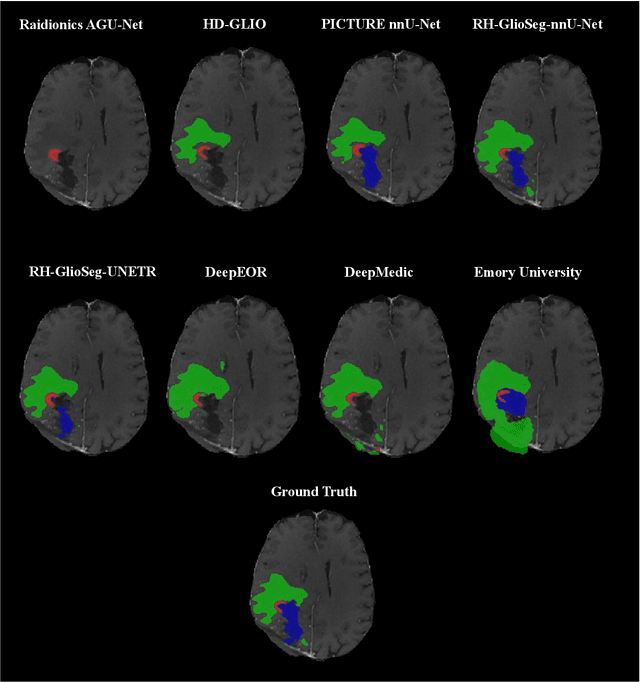
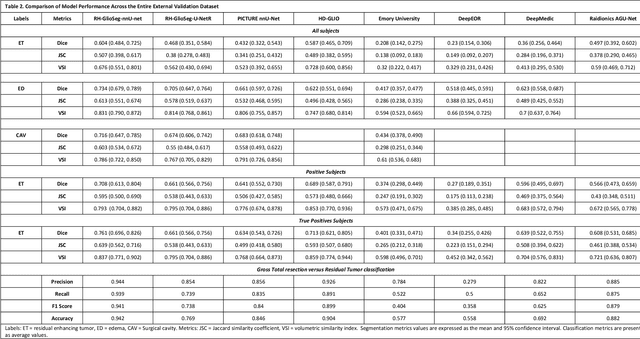
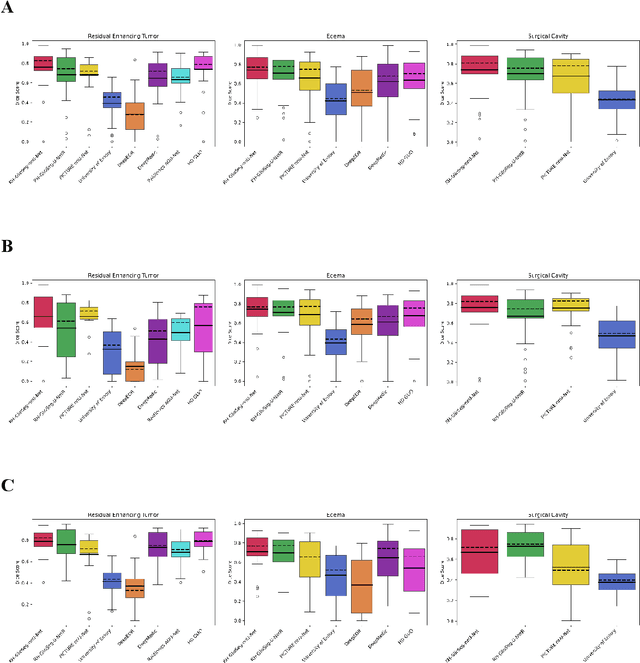
Abstract:Accurately assessing tumor removal is paramount in the management of glioblastoma. We developed a pipeline using MRI scans and neural networks to segment tumor subregions and the surgical cavity in postoperative images. Our model excels in accurately classifying the extent of resection, offering a valuable tool for clinicians in assessing treatment effectiveness.
A Fully Automated Pipeline Using Swin Transformers for Deep Learning-Based Blood Segmentation on Head CT Scans After Aneurysmal Subarachnoid Hemorrhage
Dec 29, 2023Abstract:Background: Accurate volumetric assessment of spontaneous subarachnoid hemorrhage (SAH) is a labor-intensive task performed with current manual and semiautomatic methods that might be relevant for its clinical and prognostic implications. In the present research, we sought to develop and validate an artificial intelligence-driven, fully automated blood segmentation tool for SAH patients via noncontrast computed tomography (NCCT) scans employing a transformer-based Swin UNETR architecture. Methods: We retrospectively analyzed NCCT scans from patients with confirmed aneurysmal subarachnoid hemorrhage (aSAH) utilizing the Swin UNETR for segmentation. The performance of the proposed method was evaluated against manually segmented ground truth data using metrics such as Dice score, intersection over union (IoU), the volumetric similarity index (VSI), the symmetric average surface distance (SASD), and sensitivity and specificity. A validation cohort from an external institution was included to test the generalizability of the model. Results: The model demonstrated high accuracy with robust performance metrics across the internal and external validation cohorts. Notably, it achieved high Dice coefficient (0.873), IoU (0.810), VSI (0.840), sensitivity (0.821) and specificity (0.996) values and a low SASD (1.866), suggesting proficiency in segmenting blood in SAH patients. The model's efficiency was reflected in its processing speed, indicating potential for real-time applications. Conclusions: Our Swin UNETR-based model offers significant advances in the automated segmentation of blood after aSAH on NCCT images. Despite the computational intensity, the model operates effectively on standard hardware with a user-friendly interface, facilitating broader clinical adoption. Further validation across diverse datasets is warranted to confirm its clinical reliability.
Enhanced Mortality Prediction In Patients With Subarachnoid Haemorrhage Using A Deep Learning Model Based On The Initial CT Scan
Aug 25, 2023
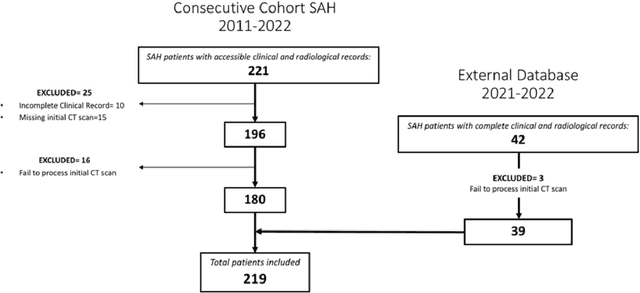
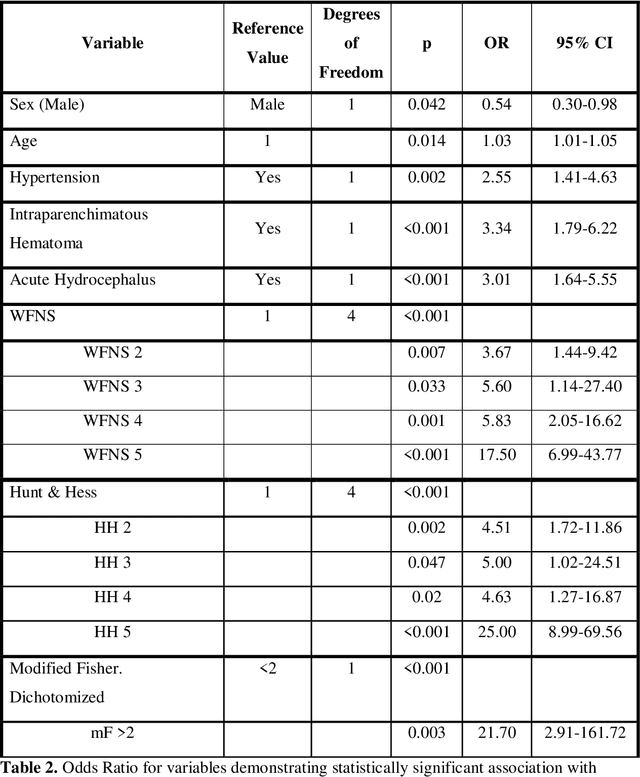
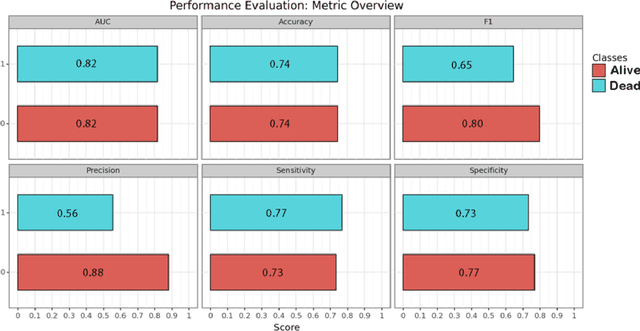
Abstract:PURPOSE: Subarachnoid hemorrhage (SAH) entails high morbidity and mortality rates. Convolutional neural networks (CNN), a form of deep learning, are capable of generating highly accurate predictions from imaging data. Our objective was to predict mortality in SAH patients by processing the initial CT scan on a CNN based algorithm. METHODS: Retrospective multicentric study of a consecutive cohort of patients with SAH between 2011-2022. Demographic, clinical and radiological variables were analyzed. Pre-processed baseline CT scan images were used as the input for training a CNN using AUCMEDI Framework. Our model's architecture leverages the DenseNet-121 structure, employing transfer learning principles. The output variable was mortality in the first three months. Performance of the model was evaluated by statistical parameters conventionally used in studies involving artificial intelligence methods. RESULTS: Images from 219 patients were processed, 175 for training and validation of the CNN and 44 for its evaluation. 52%(115/219) of patients were female, and the median age was 58(SD=13.06) years. 18.5%(39/219) were idiopathic SAH. Mortality rate was 28.5%(63/219). The model showed good accuracy at predicting mortality in SAH patients exclusively using the images of the initial CT scan (Accuracy=74%, F1=75% and AUC=82%). CONCLUSION: Modern image processing techniques based on AI and CNN make possible to predict mortality in SAH patients with high accuracy using CT scan images as the only input. These models might be optimized by including more data and patients resulting in better training, development and performance on tasks which are beyond the skills of conventional clinical knowledge.
The Rio Hortega University Hospital Glioblastoma dataset: a comprehensive collection of preoperative, early postoperative and recurrence MRI scans (RHUH-GBM)
May 02, 2023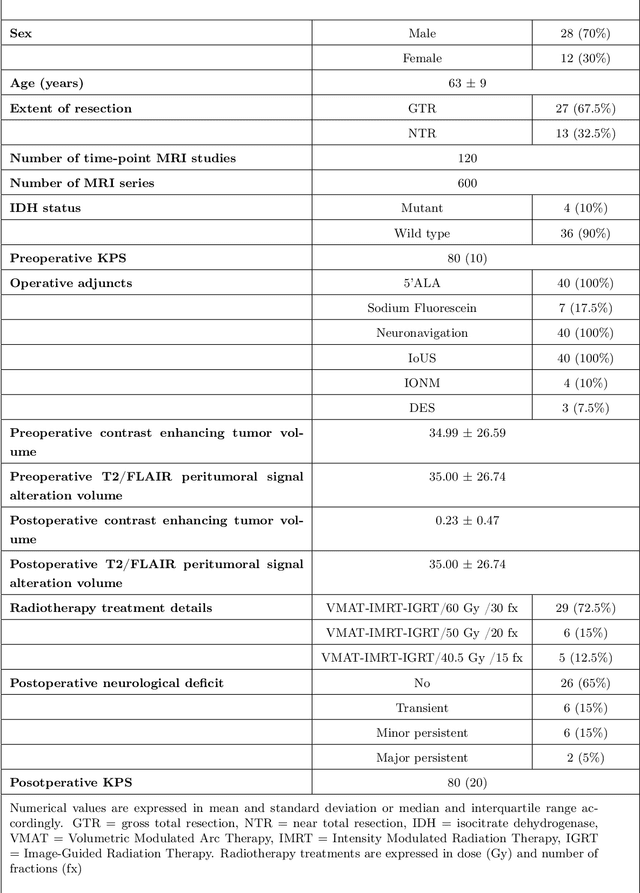
Abstract:Glioblastoma, a highly aggressive primary brain tumor, is associated with poor patient outcomes. Although magnetic resonance imaging (MRI) plays a critical role in diagnosing, characterizing, and forecasting glioblastoma progression, public MRI repositories present significant drawbacks, including insufficient postoperative and follow-up studies as well as expert tumor segmentations. To address these issues, we present the "R\'io Hortega University Hospital Glioblastoma Dataset (RHUH-GBM)," a collection of multiparametric MRI images, volumetric assessments, molecular data, and survival details for glioblastoma patients who underwent total or near-total enhancing tumor resection. The dataset features expert-corrected segmentations of tumor subregions, offering valuable ground truth data for developing algorithms for postoperative and follow-up MRI scans. The public release of the RHUH-GBM dataset significantly contributes to glioblastoma research, enabling the scientific community to study recurrence patterns and develop new diagnostic and prognostic models. This may result in more personalized, effective treatments and ultimately improved patient outcomes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge