Roman C. Maron
Clinical Melanoma Diagnosis with Artificial Intelligence: Insights from a Prospective Multicenter Study
Jan 25, 2024Abstract:Early detection of melanoma, a potentially lethal type of skin cancer with high prevalence worldwide, improves patient prognosis. In retrospective studies, artificial intelligence (AI) has proven to be helpful for enhancing melanoma detection. However, there are few prospective studies confirming these promising results. Existing studies are limited by low sample sizes, too homogenous datasets, or lack of inclusion of rare melanoma subtypes, preventing a fair and thorough evaluation of AI and its generalizability, a crucial aspect for its application in the clinical setting. Therefore, we assessed 'All Data are Ext' (ADAE), an established open-source ensemble algorithm for detecting melanomas, by comparing its diagnostic accuracy to that of dermatologists on a prospectively collected, external, heterogeneous test set comprising eight distinct hospitals, four different camera setups, rare melanoma subtypes, and special anatomical sites. We advanced the algorithm with real test-time augmentation (R-TTA, i.e. providing real photographs of lesions taken from multiple angles and averaging the predictions), and evaluated its generalization capabilities. Overall, the AI showed higher balanced accuracy than dermatologists (0.798, 95% confidence interval (CI) 0.779-0.814 vs. 0.781, 95% CI 0.760-0.802; p<0.001), obtaining a higher sensitivity (0.921, 95% CI 0.900- 0.942 vs. 0.734, 95% CI 0.701-0.770; p<0.001) at the cost of a lower specificity (0.673, 95% CI 0.641-0.702 vs. 0.828, 95% CI 0.804-0.852; p<0.001). As the algorithm exhibited a significant performance advantage on our heterogeneous dataset exclusively comprising melanoma-suspicious lesions, AI may offer the potential to support dermatologists particularly in diagnosing challenging cases.
Mitigating the Influence of Domain Shift in Skin Lesion Classification: A Benchmark Study of Unsupervised Domain Adaptation Methods on Dermoscopic Images
Oct 05, 2023



Abstract:The potential of deep neural networks in skin lesion classification has already been demonstrated to be on-par if not superior to the dermatologists diagnosis. However, the performance of these models usually deteriorates when the test data differs significantly from the training data (i.e. domain shift). This concerning limitation for models intended to be used in real-world skin lesion classification tasks poses a risk to patients. For example, different image acquisition systems or previously unseen anatomical sites on the patient can suffice to cause such domain shifts. Mitigating the negative effect of such shifts is therefore crucial, but developing effective methods to address domain shift has proven to be challenging. In this study, we carry out an in-depth analysis of eight different unsupervised domain adaptation methods to analyze their effectiveness in improving generalization for dermoscopic datasets. To ensure robustness of our findings, we test each method on a total of ten distinct datasets, thereby covering a variety of possible domain shifts. In addition, we investigated which factors in the domain shifted datasets have an impact on the effectiveness of domain adaptation methods. Our findings show that all of the eight domain adaptation methods result in improved AUPRC for the majority of analyzed datasets. Altogether, these results indicate that unsupervised domain adaptations generally lead to performance improvements for the binary melanoma-nevus classification task regardless of the nature of the domain shift. However, small or heavily imbalanced datasets lead to a reduced conformity of the results due to the influence of these factors on the methods performance.
Using Multiple Dermoscopic Photographs of One Lesion Improves Melanoma Classification via Deep Learning: A Prognostic Diagnostic Accuracy Study
Jun 05, 2023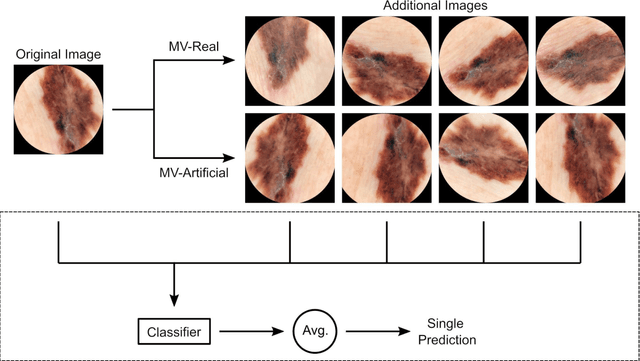
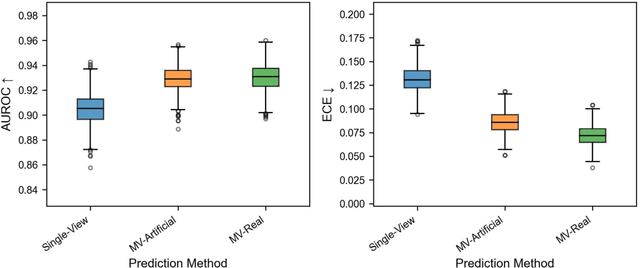
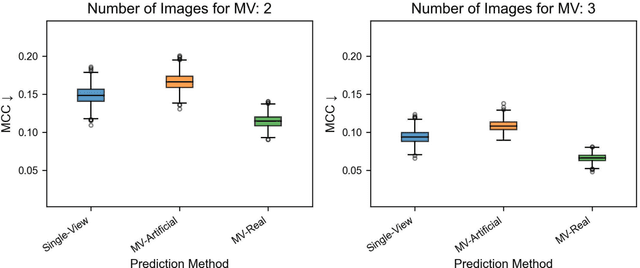
Abstract:Background: Convolutional neural network (CNN)-based melanoma classifiers face several challenges that limit their usefulness in clinical practice. Objective: To investigate the impact of multiple real-world dermoscopic views of a single lesion of interest on a CNN-based melanoma classifier. Methods: This study evaluated 656 suspected melanoma lesions. Classifier performance was measured using area under the receiver operating characteristic curve (AUROC), expected calibration error (ECE) and maximum confidence change (MCC) for (I) a single-view scenario, (II) a multiview scenario using multiple artificially modified images per lesion and (III) a multiview scenario with multiple real-world images per lesion. Results: The multiview approach with real-world images significantly increased the AUROC from 0.905 (95% CI, 0.879-0.929) in the single-view approach to 0.930 (95% CI, 0.909-0.951). ECE and MCC also improved significantly from 0.131 (95% CI, 0.105-0.159) to 0.072 (95% CI: 0.052-0.093) and from 0.149 (95% CI, 0.125-0.171) to 0.115 (95% CI: 0.099-0.131), respectively. Comparing multiview real-world to artificially modified images showed comparable diagnostic accuracy and uncertainty estimation, but significantly worse robustness for the latter. Conclusion: Using multiple real-world images is an inexpensive method to positively impact the performance of a CNN-based melanoma classifier.
Domain shifts in dermoscopic skin cancer datasets: Evaluation of essential limitations for clinical translation
Apr 18, 2023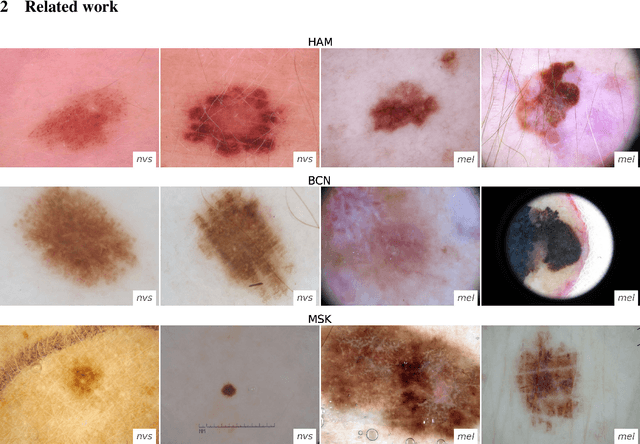
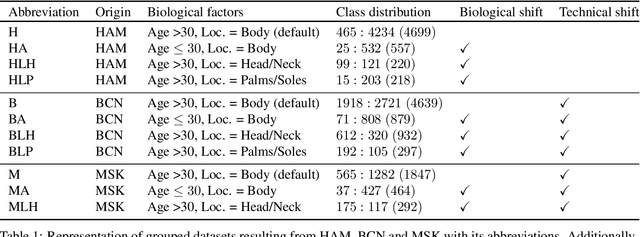
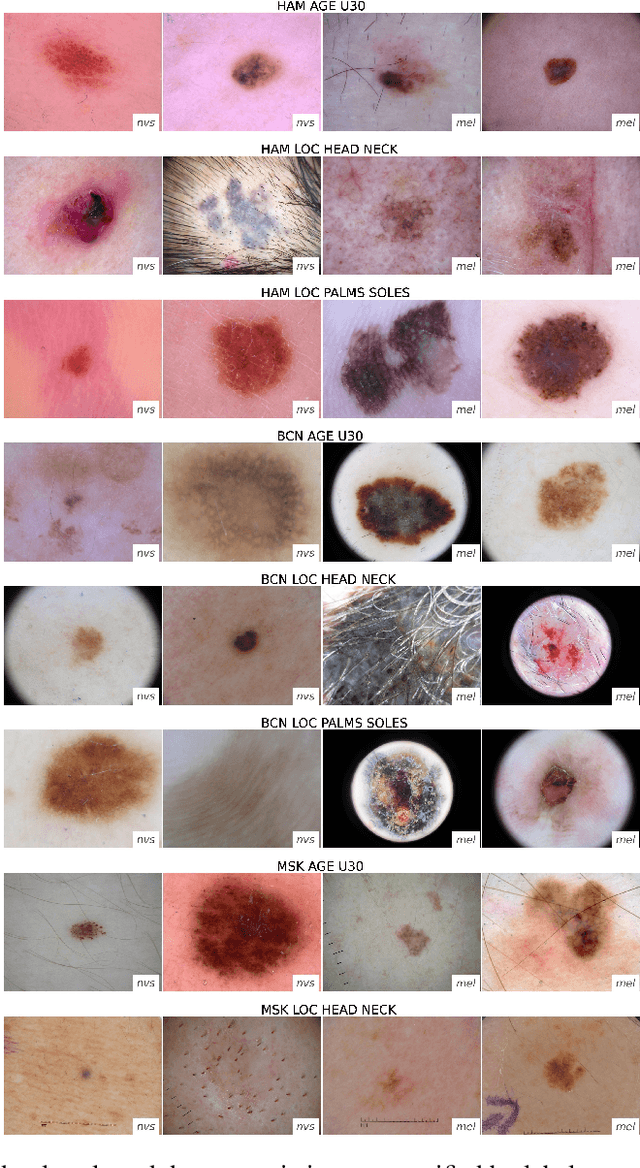
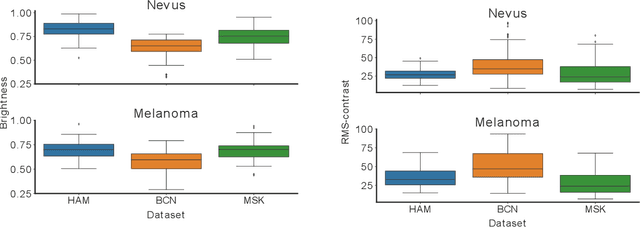
Abstract:The limited ability of Convolutional Neural Networks to generalize to images from previously unseen domains is a major limitation, in particular, for safety-critical clinical tasks such as dermoscopic skin cancer classification. In order to translate CNN-based applications into the clinic, it is essential that they are able to adapt to domain shifts. Such new conditions can arise through the use of different image acquisition systems or varying lighting conditions. In dermoscopy, shifts can also occur as a change in patient age or occurence of rare lesion localizations (e.g. palms). These are not prominently represented in most training datasets and can therefore lead to a decrease in performance. In order to verify the generalizability of classification models in real world clinical settings it is crucial to have access to data which mimics such domain shifts. To our knowledge no dermoscopic image dataset exists where such domain shifts are properly described and quantified. We therefore grouped publicly available images from ISIC archive based on their metadata (e.g. acquisition location, lesion localization, patient age) to generate meaningful domains. To verify that these domains are in fact distinct, we used multiple quantification measures to estimate the presence and intensity of domain shifts. Additionally, we analyzed the performance on these domains with and without an unsupervised domain adaptation technique. We observed that in most of our grouped domains, domain shifts in fact exist. Based on our results, we believe these datasets to be helpful for testing the generalization capabilities of dermoscopic skin cancer classifiers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge