Renate Krause
Items or Relations -- what do Artificial Neural Networks learn?
Apr 15, 2024
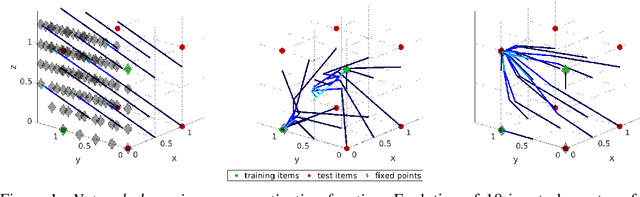

Abstract:What has an Artificial Neural Network (ANN) learned after being successfully trained to solve a task - the set of training items or the relations between them? This question is difficult to answer for modern applied ANNs because of their enormous size and complexity. Therefore, here we consider a low-dimensional network and a simple task, i.e., the network has to reproduce a set of training items identically. We construct the family of solutions analytically and use standard learning algorithms to obtain numerical solutions. These numerical solutions differ depending on the optimization algorithm and the weight initialization and are shown to be particular members of the family of analytical solutions. In this simple setting, we observe that the general structure of the network weights represents the training set's symmetry group, i.e., the relations between training items. As a consequence, linear networks generalize, i.e., reproduce items that were not part of the training set but are consistent with the symmetry of the training set. In contrast, non-linear networks tend to learn individual training items and show associative memory. At the same time, their ability to generalize is limited. A higher degree of generalization is obtained for networks whose activation function contains a linear regime, such as tanh. Our results suggest ANN's ability to generalize - instead of learning items - could be improved by generating a sufficiently big set of elementary operations to represent relations and strongly depends on the applied non-linearity.
Synaptic partner prediction from point annotations in insect brains
Jul 16, 2018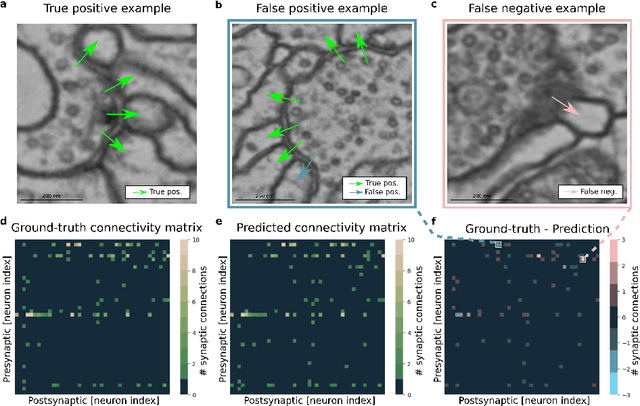
Abstract:High-throughput electron microscopy allows recording of lar- ge stacks of neural tissue with sufficient resolution to extract the wiring diagram of the underlying neural network. Current efforts to automate this process focus mainly on the segmentation of neurons. However, in order to recover a wiring diagram, synaptic partners need to be identi- fied as well. This is especially challenging in insect brains like Drosophila melanogaster, where one presynaptic site is associated with multiple post- synaptic elements. Here we propose a 3D U-Net architecture to directly identify pairs of voxels that are pre- and postsynaptic to each other. To that end, we formulate the problem of synaptic partner identification as a classification problem on long-range edges between voxels to encode both the presence of a synaptic pair and its direction. This formulation allows us to directly learn from synaptic point annotations instead of more ex- pensive voxel-based synaptic cleft or vesicle annotations. We evaluate our method on the MICCAI 2016 CREMI challenge and improve over the current state of the art, producing 3% fewer errors than the next best method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge