Julia Buhmann
Microtubule Tracking in Electron Microscopy Volumes
Sep 17, 2020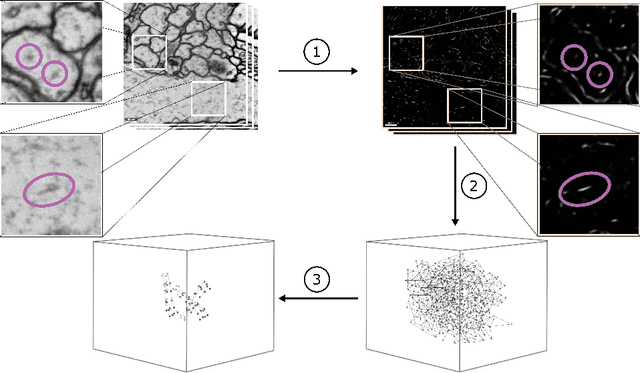
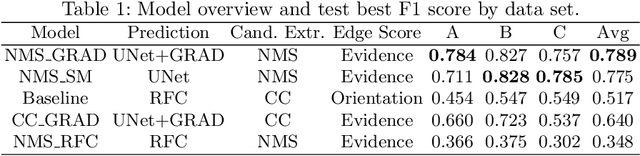
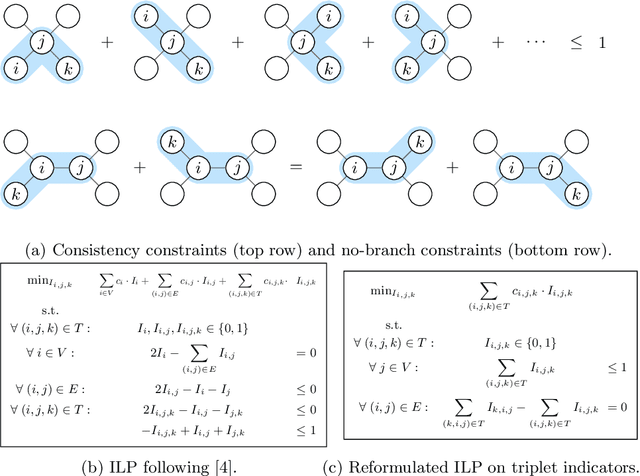
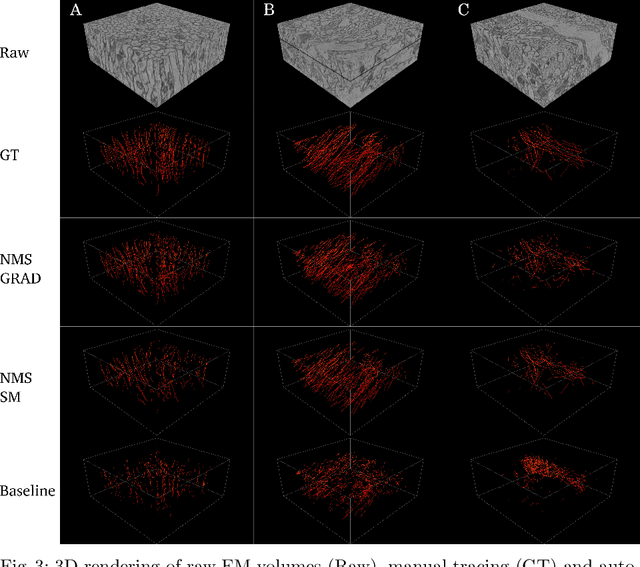
Abstract:We present a method for microtubule tracking in electron microscopy volumes. Our method first identifies a sparse set of voxels that likely belong to microtubules. Similar to prior work, we then enumerate potential edges between these voxels, which we represent in a candidate graph. Tracks of microtubules are found by selecting nodes and edges in the candidate graph by solving a constrained optimization problem incorporating biological priors on microtubule structure. For this, we present a novel integer linear programming formulation, which results in speed-ups of three orders of magnitude and an increase of 53% in accuracy compared to prior art (evaluated on three 1.2 x 4 x 4$\mu$m volumes of Drosophila neural tissue). We also propose a scheme to solve the optimization problem in a block-wise fashion, which allows distributed tracking and is necessary to process very large electron microscopy volumes. Finally, we release a benchmark dataset for microtubule tracking, here used for training, testing and validation, consisting of eight 30 x 1000 x 1000 voxel blocks (1.2 x 4 x 4$\mu$m) of densely annotated microtubules in the CREMI data set (https://github.com/nilsec/micron).
Synaptic partner prediction from point annotations in insect brains
Jul 16, 2018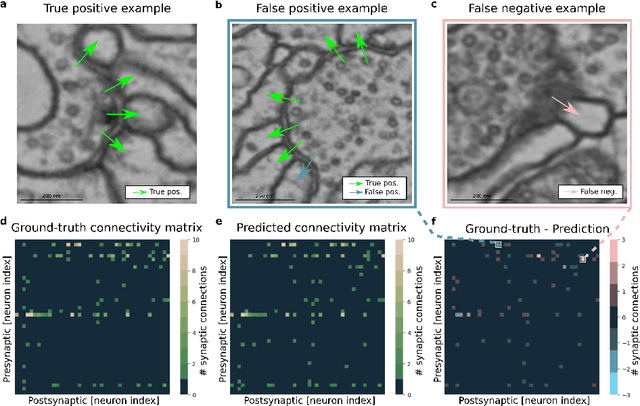
Abstract:High-throughput electron microscopy allows recording of lar- ge stacks of neural tissue with sufficient resolution to extract the wiring diagram of the underlying neural network. Current efforts to automate this process focus mainly on the segmentation of neurons. However, in order to recover a wiring diagram, synaptic partners need to be identi- fied as well. This is especially challenging in insect brains like Drosophila melanogaster, where one presynaptic site is associated with multiple post- synaptic elements. Here we propose a 3D U-Net architecture to directly identify pairs of voxels that are pre- and postsynaptic to each other. To that end, we formulate the problem of synaptic partner identification as a classification problem on long-range edges between voxels to encode both the presence of a synaptic pair and its direction. This formulation allows us to directly learn from synaptic point annotations instead of more ex- pensive voxel-based synaptic cleft or vesicle annotations. We evaluate our method on the MICCAI 2016 CREMI challenge and improve over the current state of the art, producing 3% fewer errors than the next best method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge