Rebecca Rojansky
iSight: Towards expert-AI co-assessment for improved immunohistochemistry staining interpretation
Feb 03, 2026Abstract:Immunohistochemistry (IHC) provides information on protein expression in tissue sections and is commonly used to support pathology diagnosis and disease triage. While AI models for H\&E-stained slides show promise, their applicability to IHC is limited due to domain-specific variations. Here we introduce HPA10M, a dataset that contains 10,495,672 IHC images from the Human Protein Atlas with comprehensive metadata included, and encompasses 45 normal tissue types and 20 major cancer types. Based on HPA10M, we trained iSight, a multi-task learning framework for automated IHC staining assessment. iSight combines visual features from whole-slide images with tissue metadata through a token-level attention mechanism, simultaneously predicting staining intensity, location, quantity, tissue type, and malignancy status. On held-out data, iSight achieved 85.5\% accuracy for location, 76.6\% for intensity, and 75.7\% for quantity, outperforming fine-tuned foundation models (PLIP, CONCH) by 2.5--10.2\%. In addition, iSight demonstrates well-calibrated predictions with expected calibration errors of 0.0150-0.0408. Furthermore, in a user study with eight pathologists evaluating 200 images from two datasets, iSight outperformed initial pathologist assessments on the held-out HPA dataset (79\% vs 68\% for location, 70\% vs 57\% for intensity, 68\% vs 52\% for quantity). Inter-pathologist agreement also improved after AI assistance in both held-out HPA (Cohen's $κ$ increased from 0.63 to 0.70) and Stanford TMAD datasets (from 0.74 to 0.76), suggesting expert--AI co-assessment can improve IHC interpretation. This work establishes a foundation for AI systems that can improve IHC diagnostic accuracy and highlights the potential for integrating iSight into clinical workflows to enhance the consistency and reliability of IHC assessment.
LymphoML: An interpretable artificial intelligence-based method identifies morphologic features that correlate with lymphoma subtype
Nov 20, 2023



Abstract:The accurate classification of lymphoma subtypes using hematoxylin and eosin (H&E)-stained tissue is complicated by the wide range of morphological features these cancers can exhibit. We present LymphoML - an interpretable machine learning method that identifies morphologic features that correlate with lymphoma subtypes. Our method applies steps to process H&E-stained tissue microarray cores, segment nuclei and cells, compute features encompassing morphology, texture, and architecture, and train gradient-boosted models to make diagnostic predictions. LymphoML's interpretable models, developed on a limited volume of H&E-stained tissue, achieve non-inferior diagnostic accuracy to pathologists using whole-slide images and outperform black box deep-learning on a dataset of 670 cases from Guatemala spanning 8 lymphoma subtypes. Using SHapley Additive exPlanation (SHAP) analysis, we assess the impact of each feature on model prediction and find that nuclear shape features are most discriminative for DLBCL (F1-score: 78.7%) and classical Hodgkin lymphoma (F1-score: 74.5%). Finally, we provide the first demonstration that a model combining features from H&E-stained tissue with features from a standardized panel of 6 immunostains results in a similar diagnostic accuracy (85.3%) to a 46-stain panel (86.1%).
DLBCL-Morph: Morphological features computed using deep learning for an annotated digital DLBCL image set
Sep 24, 2020


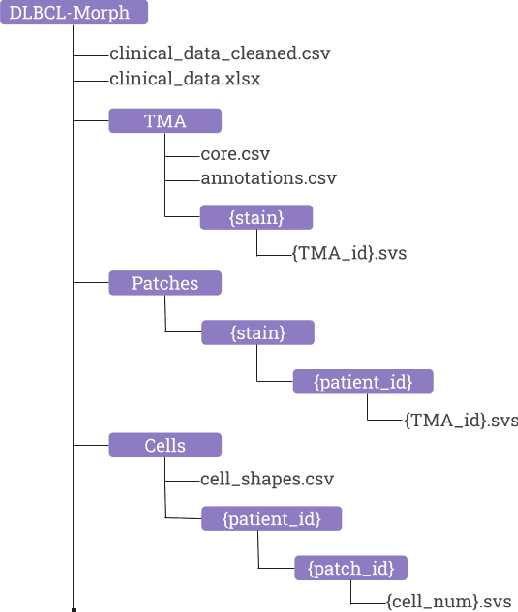
Abstract:Diffuse Large B-Cell Lymphoma (DLBCL) is the most common non-Hodgkin lymphoma. Though histologically DLBCL shows varying morphologies, no morphologic features have been consistently demonstrated to correlate with prognosis. We present a morphologic analysis of histology sections from 209 DLBCL cases with associated clinical and cytogenetic data. Duplicate tissue core sections were arranged in tissue microarrays (TMAs), and replicate sections were stained with H&E and immunohistochemical stains for CD10, BCL6, MUM1, BCL2, and MYC. The TMAs are accompanied by pathologist-annotated regions-of-interest (ROIs) that identify areas of tissue representative of DLBCL. We used a deep learning model to segment all tumor nuclei in the ROIs, and computed several geometric features for each segmented nucleus. We fit a Cox proportional hazards model to demonstrate the utility of these geometric features in predicting survival outcome, and found that it achieved a C-index (95% CI) of 0.635 (0.574,0.691). Our finding suggests that geometric features computed from tumor nuclei are of prognostic importance, and should be validated in prospective studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge