Raunak Dey
ASC-Net: Unsupervised Medical Anomaly Segmentation Using an Adversarial-based Selective Cutting Network
Dec 16, 2021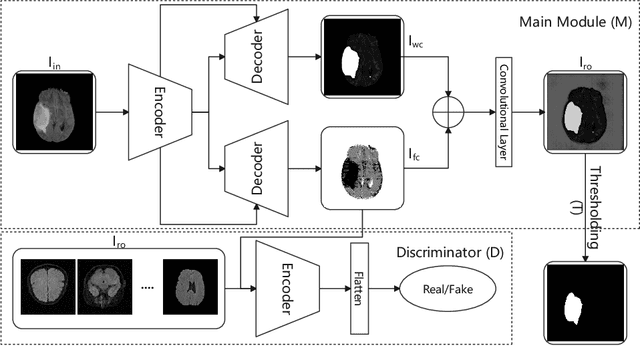
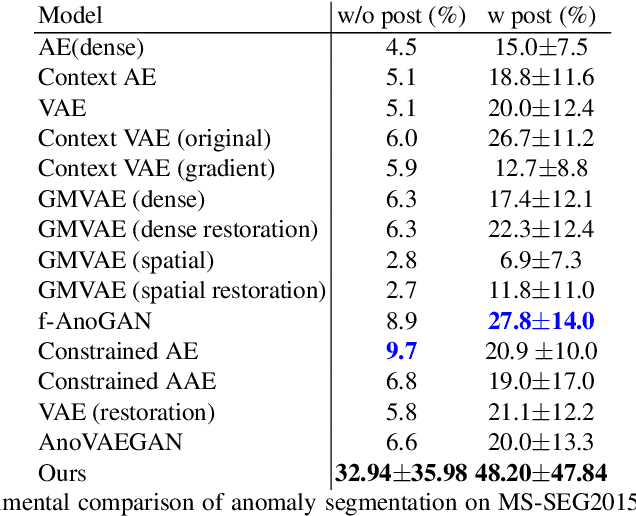
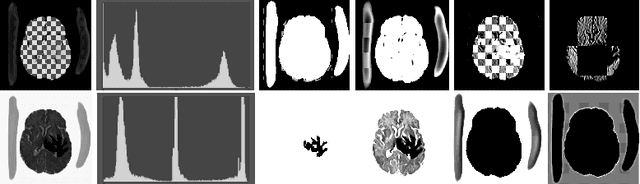
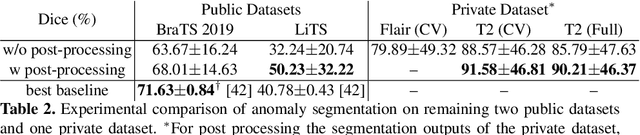
Abstract:In this paper we consider the problem of unsupervised anomaly segmentation in medical images, which has attracted increasing attention in recent years due to the expensive pixel-level annotations from experts and the existence of a large amount of unannotated normal and abnormal image scans. We introduce a segmentation network that utilizes adversarial learning to partition an image into two cuts, with one of them falling into a reference distribution provided by the user. This Adversarial-based Selective Cutting network (ASC-Net) bridges the two domains of cluster-based deep segmentation and adversarial-based anomaly/novelty detection algorithms. Our ASC-Net learns from normal and abnormal medical scans to segment anomalies in medical scans without any masks for supervision. We evaluate this unsupervised anomly segmentation model on three public datasets, i.e., BraTS 2019 for brain tumor segmentation, LiTS for liver lesion segmentation, and MS-SEG 2015 for brain lesion segmentation, and also on a private dataset for brain tumor segmentation. Compared to existing methods, our model demonstrates tremendous performance gains in unsupervised anomaly segmentation tasks. Although there is still room to further improve performance compared to supervised learning algorithms, the promising experimental results and interesting observations shed light on building an unsupervised learning algorithm for medical anomaly identification using user-defined knowledge.
ASC-Net : Adversarial-based Selective Network for Unsupervised Anomaly Segmentation
Mar 05, 2021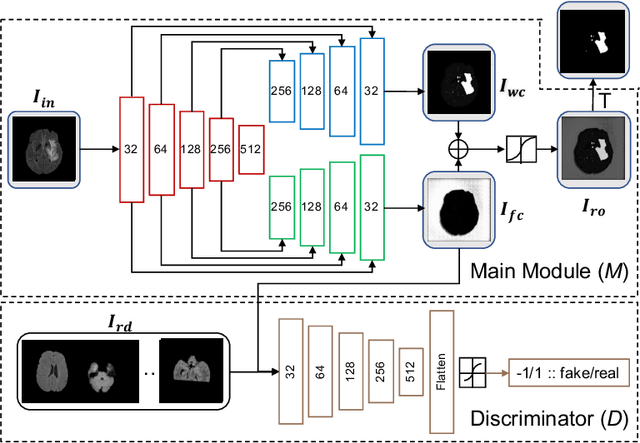
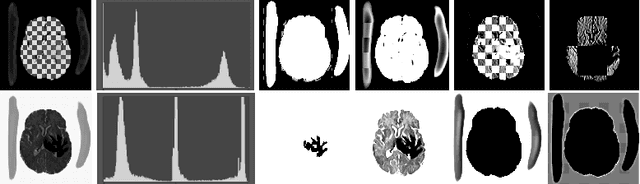

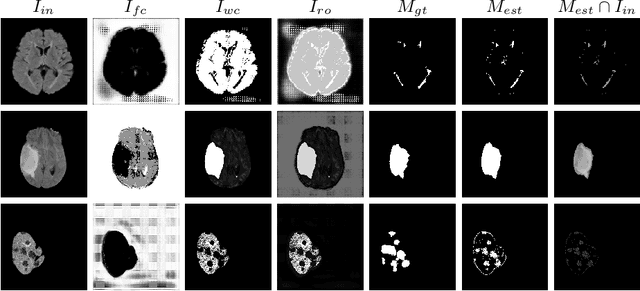
Abstract:We introduce a neural network framework, utilizing adversarial learning to partition an image into two cuts, with one cut falling into a reference distribution provided by the user. This concept tackles the task of unsupervised anomaly segmentation, which has attracted increasing attention in recent years due to their broad applications in tasks with unlabelled data. This Adversarial-based Selective Cutting network (ASC-Net) bridges the two domains of cluster-based deep learning methods and adversarial-based anomaly/novelty detection algorithms. We evaluate this unsupervised learning model on BraTS brain tumor segmentation, LiTS liver lesion segmentation, and MS-SEG2015 segmentation tasks. Compared to existing methods like the AnoGAN family, our model demonstrates tremendous performance gains in unsupervised anomaly segmentation tasks. Although there is still room to further improve performance compared to supervised learning algorithms, the promising experimental results shed light on building an unsupervised learning algorithm using user-defined knowledge.
Hybrid Cascaded Neural Network for Liver Lesion Segmentation
Oct 08, 2019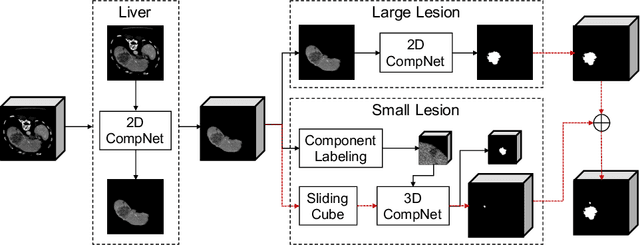
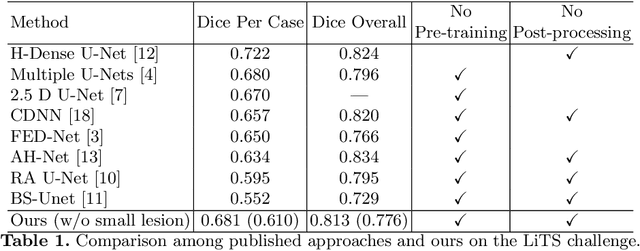
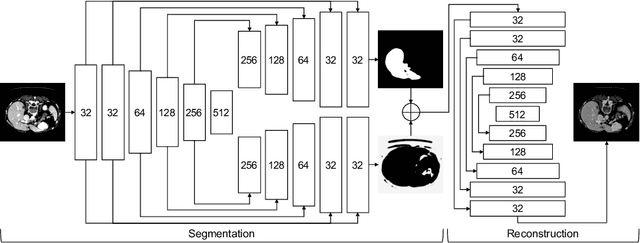
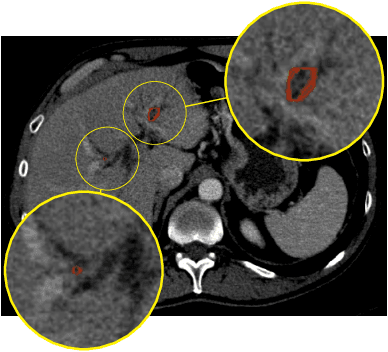
Abstract:Automatic liver lesion segmentation is a challenging task while having a significant impact on assisting medical professionals in the designing of effective treatment and planning proper care. In this paper we propose a cascaded system that combines both 2D and 3D convolutional neural networks to effectively segment hepatic lesions. Our 2D network operates on a slice by slice basis to segment the liver and larger tumors, while we use a 3D network to detect small lesions that are often missed in a 2D segmentation design. We employ this algorithm on the LiTS challenge obtaining a Dice score per case of 68.1%, which performs the best among all non pre-trained models and the second best among published methods. We also perform two-fold cross-validation to reveal the over- and under-segmentation issues in the LiTS annotations.
CompNet: Complementary Segmentation Network for Brain MRI Extraction
Jun 17, 2018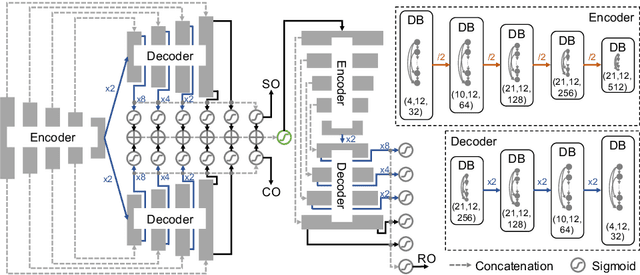
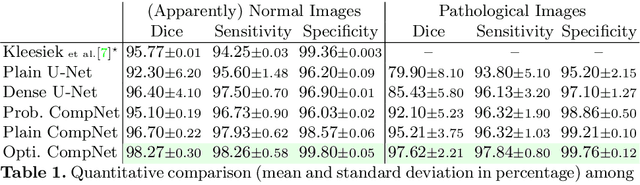
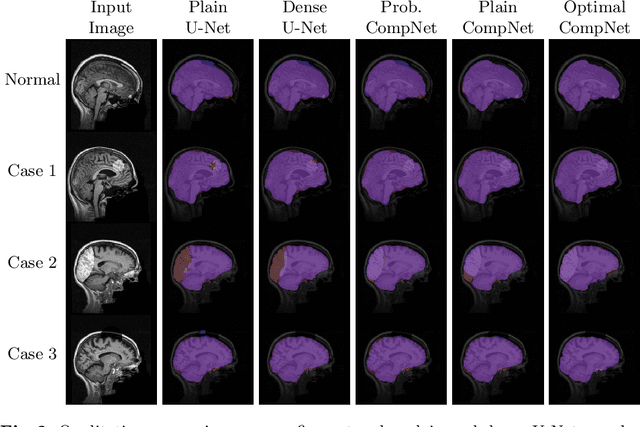
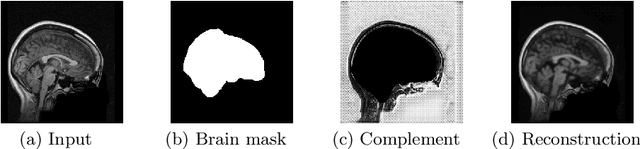
Abstract:Brain extraction is a fundamental step for most brain imaging studies. In this paper, we investigate the problem of skull stripping and propose complementary segmentation networks (CompNets) to accurately extract the brain from T1-weighted MRI scans, for both normal and pathological brain images. The proposed networks are designed in the framework of encoder-decoder networks and have two pathways to learn features from both the brain tissue and its complementary part located outside of the brain. The complementary pathway extracts the features in the non-brain region and leads to a robust solution to brain extraction from MRIs with pathologies, which do not exist in our training dataset. We demonstrate the effectiveness of our networks by evaluating them on the OASIS dataset, resulting in the state of the art performance under the two-fold cross-validation setting. Moreover, the robustness of our networks is verified by testing on images with introduced pathologies and by showing its invariance to unseen brain pathologies. In addition, our complementary network design is general and can be extended to address other image segmentation problems with better generalization.
Diagnostic Classification Of Lung Nodules Using 3D Neural Networks
Mar 19, 2018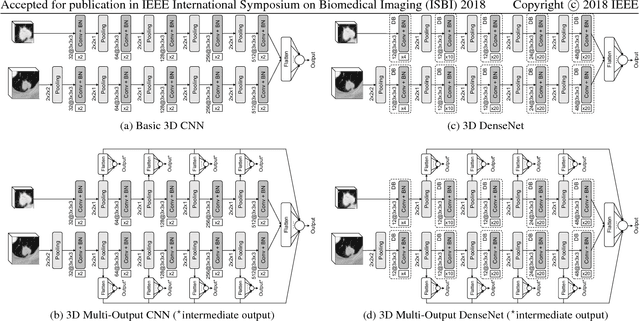
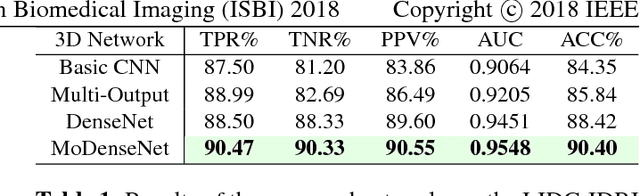
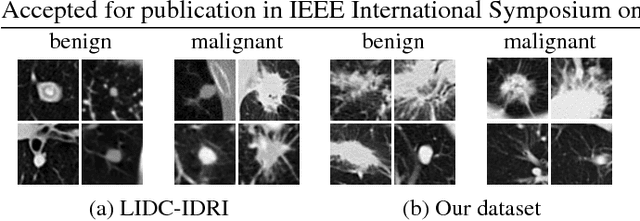
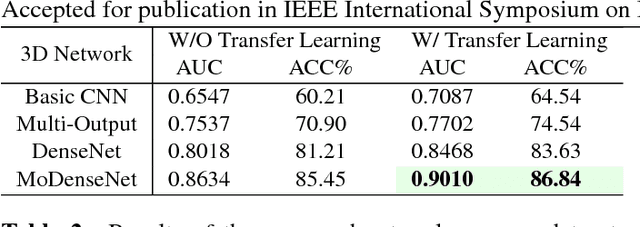
Abstract:Lung cancer is the leading cause of cancer-related death worldwide. Early diagnosis of pulmonary nodules in Computed Tomography (CT) chest scans provides an opportunity for designing effective treatment and making financial and care plans. In this paper, we consider the problem of diagnostic classification between benign and malignant lung nodules in CT images, which aims to learn a direct mapping from 3D images to class labels. To achieve this goal, four two-pathway Convolutional Neural Networks (CNN) are proposed, including a basic 3D CNN, a novel multi-output network, a 3D DenseNet, and an augmented 3D DenseNet with multi-outputs. These four networks are evaluated on the public LIDC-IDRI dataset and outperform most existing methods. In particular, the 3D multi-output DenseNet (MoDenseNet) achieves the state-of-the-art classification accuracy on the task of end-to-end lung nodule diagnosis. In addition, the networks pretrained on the LIDC-IDRI dataset can be further extended to handle smaller datasets using transfer learning. This is demonstrated on our dataset with encouraging prediction accuracy in lung nodule classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge