Raphael Ronge
When the Coffee Feature Activates on Coffins: An Analysis of Feature Extraction and Steering for Mechanistic Interpretability
Jan 06, 2026Abstract:Recent work by Anthropic on Mechanistic interpretability claims to understand and control Large Language Models by extracting human-interpretable features from their neural activation patterns using sparse autoencoders (SAEs). If successful, this approach offers one of the most promising routes for human oversight in AI safety. We conduct an initial stress-test of these claims by replicating their main results with open-source SAEs for Llama 3.1. While we successfully reproduce basic feature extraction and steering capabilities, our investigation suggests that major caution is warranted regarding the generalizability of these claims. We find that feature steering exhibits substantial fragility, with sensitivity to layer selection, steering magnitude, and context. We observe non-standard activation behavior and demonstrate the difficulty to distinguish thematically similar features from one another. While SAE-based interpretability produces compelling demonstrations in selected cases, current methods often fall short of the systematic reliability required for safety-critical applications. This suggests a necessary shift in focus from prioritizing interpretability of internal representations toward reliable prediction and control of model output. Our work contributes to a more nuanced understanding of what mechanistic interpretability has achieved and highlights fundamental challenges for AI safety that remain unresolved.
Alzheimer's Disease Diagnosis via Deep Factorization Machine Models
Aug 12, 2021
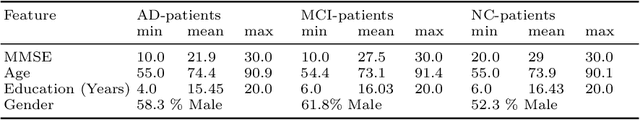


Abstract:The current state-of-the-art deep neural networks (DNNs) for Alzheimer's Disease diagnosis use different biomarker combinations to classify patients, but do not allow extracting knowledge about the interactions of biomarkers. However, to improve our understanding of the disease, it is paramount to extract such knowledge from the learned model. In this paper, we propose a Deep Factorization Machine model that combines the ability of DNNs to learn complex relationships and the ease of interpretability of a linear model. The proposed model has three parts: (i) an embedding layer to deal with sparse categorical data, (ii) a Factorization Machine to efficiently learn pairwise interactions, and (iii) a DNN to implicitly model higher order interactions. In our experiments on data from the Alzheimer's Disease Neuroimaging Initiative, we demonstrate that our proposed model classifies cognitive normal, mild cognitive impaired, and demented patients more accurately than competing models. In addition, we show that valuable knowledge about the interactions among biomarkers can be obtained.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge