Rafał Karczewski
Spacetime Geometry of Denoising in Diffusion Models
May 23, 2025Abstract:We present a novel perspective on diffusion models using the framework of information geometry. We show that the set of noisy samples, taken across all noise levels simultaneously, forms a statistical manifold -- a family of denoising probability distributions. Interpreting the noise level as a temporal parameter, we refer to this manifold as spacetime. This manifold naturally carries a Fisher-Rao metric, which defines geodesics -- shortest paths between noisy points. Notably, this family of distributions is exponential, enabling efficient geodesic computation even in high-dimensional settings without retraining or fine-tuning. We demonstrate the practical value of this geometric viewpoint in transition path sampling, where spacetime geodesics define smooth sequences of Boltzmann distributions, enabling the generation of continuous trajectories between low-energy metastable states. Code is available at: https://github.com/Aalto-QuML/diffusion-spacetime-geometry.
Devil is in the Details: Density Guidance for Detail-Aware Generation with Flow Models
Feb 09, 2025



Abstract:Diffusion models have emerged as a powerful class of generative models, capable of producing high-quality images by mapping noise to a data distribution. However, recent findings suggest that image likelihood does not align with perceptual quality: high-likelihood samples tend to be smooth, while lower-likelihood ones are more detailed. Controlling sample density is thus crucial for balancing realism and detail. In this paper, we analyze an existing technique, Prior Guidance, which scales the latent code to influence image detail. We introduce score alignment, a condition that explains why this method works and show that it can be tractably checked for any continuous normalizing flow model. We then propose Density Guidance, a principled modification of the generative ODE that enables exact log-density control during sampling. Finally, we extend Density Guidance to stochastic sampling, ensuring precise log-density control while allowing controlled variation in structure or fine details. Our experiments demonstrate that these techniques provide fine-grained control over image detail without compromising sample quality.
Diffusion Models as Cartoonists! The Curious Case of High Density Regions
Nov 02, 2024Abstract:We investigate what kind of images lie in the high-density regions of diffusion models. We introduce a theoretical mode-tracking process capable of pinpointing the exact mode of the denoising distribution, and we propose a practical high-probability sampler that consistently generates images of higher likelihood than usual samplers. Our empirical findings reveal the existence of significantly higher likelihood samples that typical samplers do not produce, often manifesting as cartoon-like drawings or blurry images depending on the noise level. Curiously, these patterns emerge in datasets devoid of such examples. We also present a novel approach to track sample likelihoods in diffusion SDEs, which remarkably incurs no additional computational cost.
What Ails Generative Structure-based Drug Design: Too Little or Too Much Expressivity?
Aug 12, 2024



Abstract:Several generative models with elaborate training and sampling procedures have been proposed recently to accelerate structure-based drug design (SBDD); however, perplexingly, their empirical performance turns out to be suboptimal. We seek to better understand this phenomenon from both theoretical and empirical perspectives. Since most of these models apply graph neural networks (GNNs), one may suspect that they inherit the representational limitations of GNNs. We analyze this aspect, establishing the first such results for protein-ligand complexes. A plausible counterview may attribute the underperformance of these models to their excessive parameterizations, inducing expressivity at the expense of generalization. We also investigate this possibility with a simple metric-aware approach that learns an economical surrogate for affinity to infer an unlabelled molecular graph and optimizes for labels conditioned on this graph and molecular properties. The resulting model achieves state-of-the-art results using 100x fewer trainable parameters and affords up to 1000x speedup. Collectively, our findings underscore the need to reassess and redirect the existing paradigm and efforts for SBDD.
Inhibited Softmax for Uncertainty Estimation in Neural Networks
Oct 03, 2018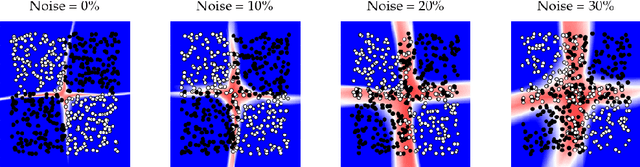
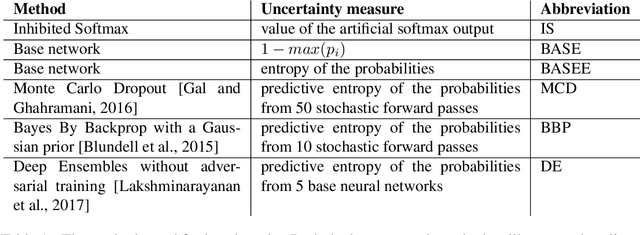
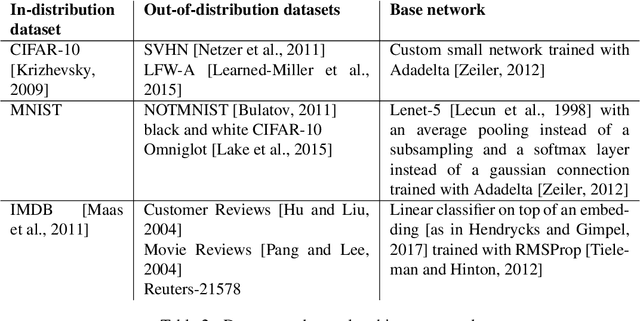
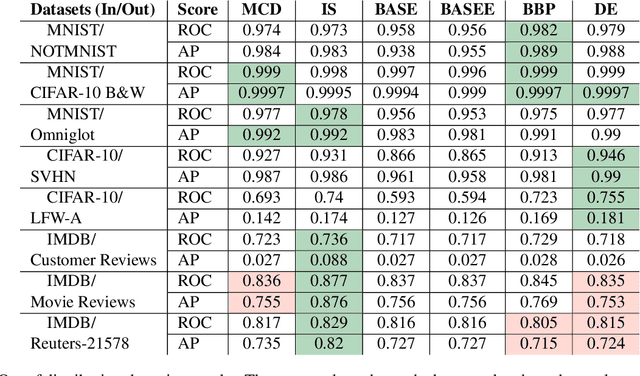
Abstract:We present a new method for uncertainty estimation and out-of-distribution detection in neural networks with softmax output. We extend softmax layer with an additional constant input. The corresponding additional output is able to represent the uncertainty of the network. The proposed method requires neither additional parameters nor multiple forward passes nor input preprocessing nor out-of-distribution datasets. We show that our method performs comparably to more computationally expensive methods and outperforms baselines on our experiments from image recognition and sentiment analysis domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge