Radim Burget
Comparing Normalization Methods for Limited Batch Size Segmentation Neural Networks
Nov 23, 2020



Abstract:The widespread use of Batch Normalization has enabled training deeper neural networks with more stable and faster results. However, the Batch Normalization works best using large batch size during training and as the state-of-the-art segmentation convolutional neural network architectures are very memory demanding, large batch size is often impossible to achieve on current hardware. We evaluate the alternative normalization methods proposed to solve this issue on a problem of binary spine segmentation from 3D CT scan. Our results show the effectiveness of Instance Normalization in the limited batch size neural network training environment. Out of all the compared methods the Instance Normalization achieved the highest result with Dice coefficient = 0.96 which is comparable to our previous results achieved by deeper network with longer training time. We also show that the Instance Normalization implementation used in this experiment is computational time efficient when compared to the network without any normalization method.
Planar 3D Transfer Learning for End to End Unimodal MRI Unbalanced Data Segmentation
Nov 23, 2020



Abstract:We present a novel approach of 2D to 3D transfer learning based on mapping pre-trained 2D convolutional neural network weights into planar 3D kernels. The method is validated by the proposed planar 3D res-u-net network with encoder transferred from the 2D VGG-16, which is applied for a single-stage unbalanced 3D image data segmentation. In particular, we evaluate the method on the MICCAI 2016 MS lesion segmentation challenge dataset utilizing solely fluid-attenuated inversion recovery (FLAIR) sequence without brain extraction for training and inference to simulate real medical praxis. The planar 3D res-u-net network performed the best both in sensitivity and Dice score amongst end to end methods processing raw MRI scans and achieved comparable Dice score to a state-of-the-art unimodal not end to end approach. Complete source code was released under the open-source license, and this paper complies with the Machine learning reproducibility checklist. By implementing practical transfer learning for 3D data representation, we could segment heavily unbalanced data without selective sampling and achieved more reliable results using less training data in a single modality. From a medical perspective, the unimodal approach gives an advantage in real praxis as it does not require co-registration nor additional scanning time during an examination. Although modern medical imaging methods capture high-resolution 3D anatomy scans suitable for computer-aided detection system processing, deployment of automatic systems for interpretation of radiology imaging is still rather theoretical in many medical areas. Our work aims to bridge the gap by offering a solution for partial research questions.
Towards Robust Voice Pathology Detection
Jul 13, 2019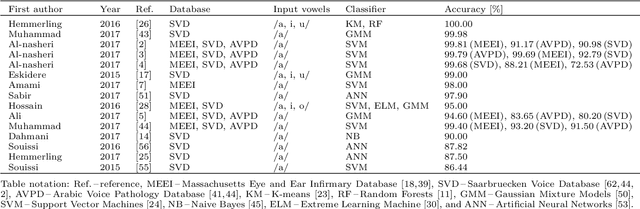
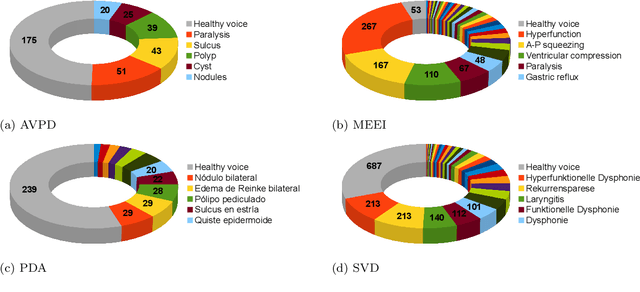
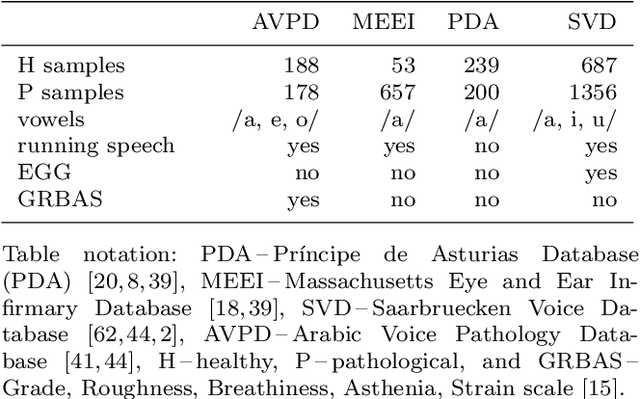
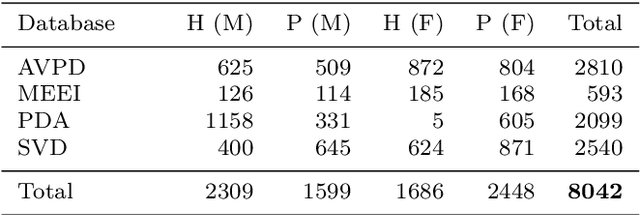
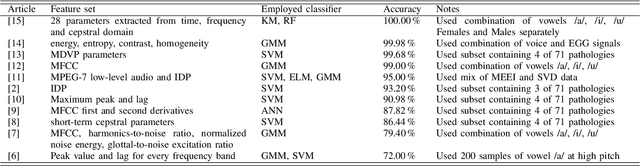
Abstract:Automatic objective non-invasive detection of pathological voice based on computerized analysis of acoustic signals can play an important role in early diagnosis, progression tracking and even effective treatment of pathological voices. In search towards such a robust voice pathology detection system we investigated 3 distinct classifiers within supervised learning and anomaly detection paradigms. We conducted a set of experiments using a variety of input data such as raw waveforms, spectrograms, mel-frequency cepstral coefficients (MFCC) and conventional acoustic (dysphonic) features (AF). In comparison with previously published works, this article is the first to utilize combination of 4 different databases comprising normophonic and pathological recordings of sustained phonation of the vowel /a/ unrestricted to a subset of vocal pathologies. Furthermore, to our best knowledge, this article is the first to explore gradient boosted trees and deep learning for this application. The following best classification performances measured by F1 score on dedicated test set were achieved: XGBoost (0.733) using AF and MFCC, DenseNet (0.621) using MFCC, and Isolation Forest (0.610) using AF. Even though these results are of exploratory character, conducted experiments do show promising potential of gradient boosting and deep learning methods to robustly detect voice pathologies.
* 11 pages, 1 figure, 10 tables. Keywords: Voice pathology detection, deep learning, gradient boosting, anomaly detection
Voice Pathology Detection Using Deep Learning: a Preliminary Study
Jul 12, 2019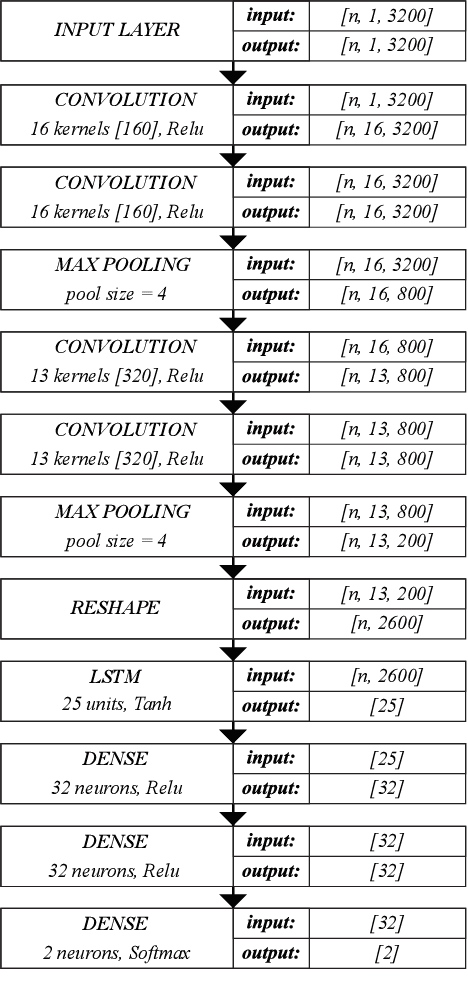


Abstract:This paper describes a preliminary investigation of Voice Pathology Detection using Deep Neural Networks (DNN). We used voice recordings of sustained vowel /a/ produced at normal pitch from German corpus Saarbruecken Voice Database (SVD). This corpus contains voice recordings and electroglottograph signals of more than 2 000 speakers. The idea behind this experiment is the use of convolutional layers in combination with recurrent Long-Short-Term-Memory (LSTM) layers on raw audio signal. Each recording was split into 64 ms Hamming windowed segments with 30 ms overlap. Our trained model achieved 71.36% accuracy with 65.04% sensitivity and 77.67% specificity on 206 validation files and 68.08% accuracy with 66.75% sensitivity and 77.89% specificity on 874 testing files. This is a promising result in favor of this approach because it is comparable to similar previously published experiment that used different methodology. Further investigation is needed to achieve the state-of-the-art results.
* 4 pages, 1 figure, 5 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge