Piotr Wójcik
UnHype: CLIP-Guided Hypernetworks for Dynamic LoRA Unlearning
Feb 03, 2026Abstract:Recent advances in large-scale diffusion models have intensified concerns about their potential misuse, particularly in generating realistic yet harmful or socially disruptive content. This challenge has spurred growing interest in effective machine unlearning, the process of selectively removing specific knowledge or concepts from a model without compromising its overall generative capabilities. Among various approaches, Low-Rank Adaptation (LoRA) has emerged as an effective and efficient method for fine-tuning models toward targeted unlearning. However, LoRA-based methods often exhibit limited adaptability to concept semantics and struggle to balance removing closely related concepts with maintaining generalization across broader meanings. Moreover, these methods face scalability challenges when multiple concepts must be erased simultaneously. To address these limitations, we introduce UnHype, a framework that incorporates hypernetworks into single- and multi-concept LoRA training. The proposed architecture can be directly plugged into Stable Diffusion as well as modern flow-based text-to-image models, where it demonstrates stable training behavior and effective concept control. During inference, the hypernetwork dynamically generates adaptive LoRA weights based on the CLIP embedding, enabling more context-aware, scalable unlearning. We evaluate UnHype across several challenging tasks, including object erasure, celebrity erasure, and explicit content removal, demonstrating its effectiveness and versatility. Repository: https://github.com/gmum/UnHype.
Token Recycling for Efficient Sequential Inference with Vision Transformers
Nov 26, 2023
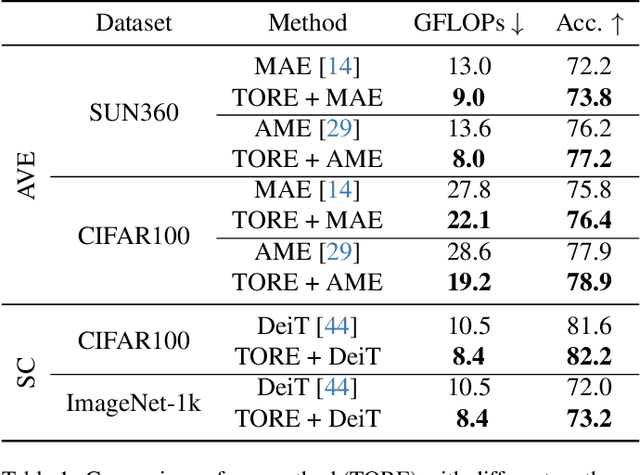
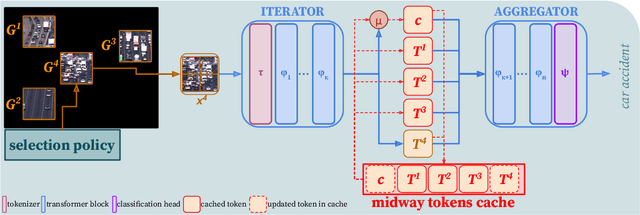
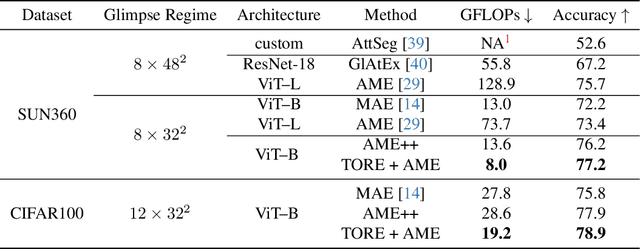
Abstract:Vision Transformers (ViTs) overpass Convolutional Neural Networks in processing incomplete inputs because they do not require the imputation of missing values. Therefore, ViTs are well suited for sequential decision-making, e.g. in the Active Visual Exploration problem. However, they are computationally inefficient because they perform a full forward pass each time a piece of new sequential information arrives. To reduce this computational inefficiency, we introduce the TOken REcycling (TORE) modification for the ViT inference, which can be used with any architecture. TORE divides ViT into two parts, iterator and aggregator. An iterator processes sequential information separately into midway tokens, which are cached. The aggregator processes midway tokens jointly to obtain the prediction. This way, we can reuse the results of computations made by iterator. Except for efficient sequential inference, we propose a complementary training policy, which significantly reduces the computational burden associated with sequential decision-making while achieving state-of-the-art accuracy.
Learning Nuclei Representations with Masked Image Modelling
Jun 29, 2023Abstract:Masked image modelling (MIM) is a powerful self-supervised representation learning paradigm, whose potential has not been widely demonstrated in medical image analysis. In this work, we show the capacity of MIM to capture rich semantic representations of Haemotoxylin & Eosin (H&E)-stained images at the nuclear level. Inspired by Bidirectional Encoder representation from Image Transformers (BEiT), we split the images into smaller patches and generate corresponding discrete visual tokens. In addition to the regular grid-based patches, typically used in visual Transformers, we introduce patches of individual cell nuclei. We propose positional encoding of the irregular distribution of these structures within an image. We pre-train the model in a self-supervised manner on H&E-stained whole-slide images of diffuse large B-cell lymphoma, where cell nuclei have been segmented. The pre-training objective is to recover the original discrete visual tokens of the masked image on the one hand, and to reconstruct the visual tokens of the masked object instances on the other. Coupling these two pre-training tasks allows us to build powerful, context-aware representations of nuclei. Our model generalizes well and can be fine-tuned on downstream classification tasks, achieving improved cell classification accuracy on PanNuke dataset by more than 5% compared to current instance segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge