Phi Nguyen Van
Echocardiography video synthesis from end diastolic semantic map via diffusion model
Oct 11, 2023Abstract:Denoising Diffusion Probabilistic Models (DDPMs) have demonstrated significant achievements in various image and video generation tasks, including the domain of medical imaging. However, generating echocardiography videos based on semantic anatomical information remains an unexplored area of research. This is mostly due to the constraints imposed by the currently available datasets, which lack sufficient scale and comprehensive frame-wise annotations for every cardiac cycle. This paper aims to tackle the aforementioned challenges by expanding upon existing video diffusion models for the purpose of cardiac video synthesis. More specifically, our focus lies in generating video using semantic maps of the initial frame during the cardiac cycle, commonly referred to as end diastole. To further improve the synthesis process, we integrate spatial adaptive normalization into multiscale feature maps. This enables the inclusion of semantic guidance during synthesis, resulting in enhanced realism and coherence of the resultant video sequences. Experiments are conducted on the CAMUS dataset, which is a highly used dataset in the field of echocardiography. Our model exhibits better performance compared to the standard diffusion technique in terms of multiple metrics, including FID, FVD, and SSMI.
Echocardiography Segmentation Using Neural ODE-based Diffeomorphic Registration Field
Jun 16, 2023Abstract:Convolutional neural networks (CNNs) have recently proven their excellent ability to segment 2D cardiac ultrasound images. However, the majority of attempts to perform full-sequence segmentation of cardiac ultrasound videos either rely on models trained only on keyframe images or fail to maintain the topology over time. To address these issues, in this work, we consider segmentation of ultrasound video as a registration estimation problem and present a novel method for diffeomorphic image registration using neural ordinary differential equations (Neural ODE). In particular, we consider the registration field vector field between frames as a continuous trajectory ODE. The estimated registration field is then applied to the segmentation mask of the first frame to obtain a segment for the whole cardiac cycle. The proposed method, Echo-ODE, introduces several key improvements compared to the previous state-of-the-art. Firstly, by solving a continuous ODE, the proposed method achieves smoother segmentation, preserving the topology of segmentation maps over the whole sequence (Hausdorff distance: 3.7-4.4). Secondly, it maintains temporal consistency between frames without explicitly optimizing for temporal consistency attributes, achieving temporal consistency in 91% of the videos in the dataset. Lastly, the proposed method is able to maintain the clinical accuracy of the segmentation maps (MAE of the LVEF: 2.7-3.1). The results show that our method surpasses the previous state-of-the-art in multiple aspects, demonstrating the importance of spatial-temporal data processing for the implementation of Neural ODEs in medical imaging applications. These findings open up new research directions for solving echocardiography segmentation tasks.
ViWOZ: A Multi-Domain Task-Oriented Dialogue Systems Dataset For Low-resource Language
Mar 15, 2022

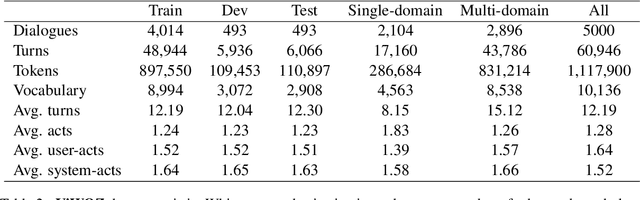

Abstract:Most of the current task-oriented dialogue systems (ToD), despite having interesting results, are designed for a handful of languages like Chinese and English. Therefore, their performance in low-resource languages is still a significant problem due to the absence of a standard dataset and evaluation policy. To address this problem, we proposed ViWOZ, a fully-annotated Vietnamese task-oriented dialogue dataset. ViWOZ is the first multi-turn, multi-domain tasked oriented dataset in Vietnamese, a low-resource language. The dataset consists of a total of 5,000 dialogues, including 60,946 fully annotated utterances. Furthermore, we provide a comprehensive benchmark of both modular and end-to-end models in low-resource language scenarios. With those characteristics, the ViWOZ dataset enables future studies on creating a multilingual task-oriented dialogue system.
Fully Automated Machine Learning Pipeline for Echocardiogram Segmentation
Jul 18, 2021

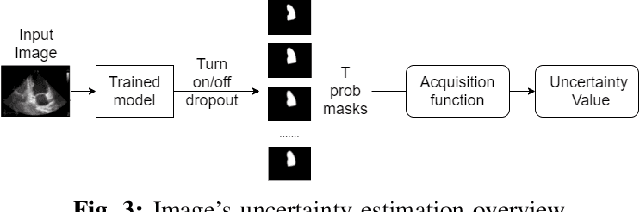

Abstract:Nowadays, cardiac diagnosis largely depends on left ventricular function assessment. With the help of the segmentation deep learning model, the assessment of the left ventricle becomes more accessible and accurate. However, deep learning technique still faces two main obstacles: the difficulty in acquiring sufficient training data and time-consuming in developing quality models. In the ordinary data acquisition process, the dataset was selected randomly from a large pool of unlabeled images for labeling, leading to massive labor time to annotate those images. Besides that, hand-designed model development is laborious and also costly. This paper introduces a pipeline that relies on Active Learning to ease the labeling work and utilizes Neural Architecture Search's idea to design the adequate deep learning model automatically. We called this Fully automated machine learning pipeline for echocardiogram segmentation. The experiment results show that our method obtained the same IOU accuracy with only two-fifths of the original training dataset, and the searched model got the same accuracy as the hand-designed model given the same training dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge