Pedro L. C. Rodrigues
University of Paris-Saclay
CP4SBI: Local Conformal Calibration of Credible Sets in Simulation-Based Inference
Aug 23, 2025Abstract:Current experimental scientists have been increasingly relying on simulation-based inference (SBI) to invert complex non-linear models with intractable likelihoods. However, posterior approximations obtained with SBI are often miscalibrated, causing credible regions to undercover true parameters. We develop $\texttt{CP4SBI}$, a model-agnostic conformal calibration framework that constructs credible sets with local Bayesian coverage. Our two proposed variants, namely local calibration via regression trees and CDF-based calibration, enable finite-sample local coverage guarantees for any scoring function, including HPD, symmetric, and quantile-based regions. Experiments on widely used SBI benchmarks demonstrate that our approach improves the quality of uncertainty quantification for neural posterior estimators using both normalizing flows and score-diffusion modeling.
Diffusion posterior sampling for simulation-based inference in tall data settings
Apr 11, 2024Abstract:Determining which parameters of a non-linear model could best describe a set of experimental data is a fundamental problem in science and it has gained much traction lately with the rise of complex large-scale simulators (a.k.a. black-box simulators). The likelihood of such models is typically intractable, which is why classical MCMC methods can not be used. Simulation-based inference (SBI) stands out in this context by only requiring a dataset of simulations to train deep generative models capable of approximating the posterior distribution that relates input parameters to a given observation. In this work, we consider a tall data extension in which multiple observations are available and one wishes to leverage their shared information to better infer the parameters of the model. The method we propose is built upon recent developments from the flourishing score-based diffusion literature and allows us to estimate the tall data posterior distribution simply using information from the score network trained on individual observations. We compare our method to recently proposed competing approaches on various numerical experiments and demonstrate its superiority in terms of numerical stability and computational cost.
Fast, accurate and lightweight sequential simulation-based inference using Gaussian locally linear mappings
Mar 16, 2024Abstract:Bayesian inference for complex models with an intractable likelihood can be tackled using algorithms performing many calls to computer simulators. These approaches are collectively known as "simulation-based inference" (SBI). Recent SBI methods have made use of neural networks (NN) to provide approximate, yet expressive constructs for the unavailable likelihood function and the posterior distribution. However, they do not generally achieve an optimal trade-off between accuracy and computational demand. In this work, we propose an alternative that provides both approximations to the likelihood and the posterior distribution, using structured mixtures of probability distributions. Our approach produces accurate posterior inference when compared to state-of-the-art NN-based SBI methods, while exhibiting a much smaller computational footprint. We illustrate our results on several benchmark models from the SBI literature.
L-C2ST: Local Diagnostics for Posterior Approximations in Simulation-Based Inference
Jun 06, 2023Abstract:Many recent works in simulation-based inference (SBI) rely on deep generative models to approximate complex, high-dimensional posterior distributions. However, evaluating whether or not these approximations can be trusted remains a challenge. Most approaches evaluate the posterior estimator only in expectation over the observation space. This limits their interpretability and is not sufficient to identify for which observations the approximation can be trusted or should be improved. Building upon the well-known classifier two-sample test (C2ST), we introduce L-C2ST, a new method that allows for a local evaluation of the posterior estimator at any given observation. It offers theoretically grounded and easy to interpret - e.g. graphical - diagnostics, and unlike C2ST, does not require access to samples from the true posterior. In the case of normalizing flow-based posterior estimators, L-C2ST can be specialized to offer better statistical power, while being computationally more efficient. On standard SBI benchmarks, L-C2ST provides comparable results to C2ST and outperforms alternative local approaches such as coverage tests based on highest predictive density (HPD). We further highlight the importance of local evaluation and the benefit of interpretability of L-C2ST on a challenging application from computational neuroscience.
Validation Diagnostics for SBI algorithms based on Normalizing Flows
Nov 24, 2022Abstract:Building on the recent trend of new deep generative models known as Normalizing Flows (NF), simulation-based inference (SBI) algorithms can now efficiently accommodate arbitrary complex and high-dimensional data distributions. The development of appropriate validation methods however has fallen behind. Indeed, most of the existing metrics either require access to the true posterior distribution, or fail to provide theoretical guarantees on the consistency of the inferred approximation beyond the one-dimensional setting. This work proposes easy to interpret validation diagnostics for multi-dimensional conditional (posterior) density estimators based on NF. It also offers theoretical guarantees based on results of local consistency. The proposed workflow can be used to check, analyse and guarantee consistent behavior of the estimator. The method is illustrated with a challenging example that involves tightly coupled parameters in the context of computational neuroscience. This work should help the design of better specified models or drive the development of novel SBI-algorithms, hence allowing to build up trust on their ability to address important questions in experimental science.
Leveraging Global Parameters for Flow-based Neural Posterior Estimation
Feb 12, 2021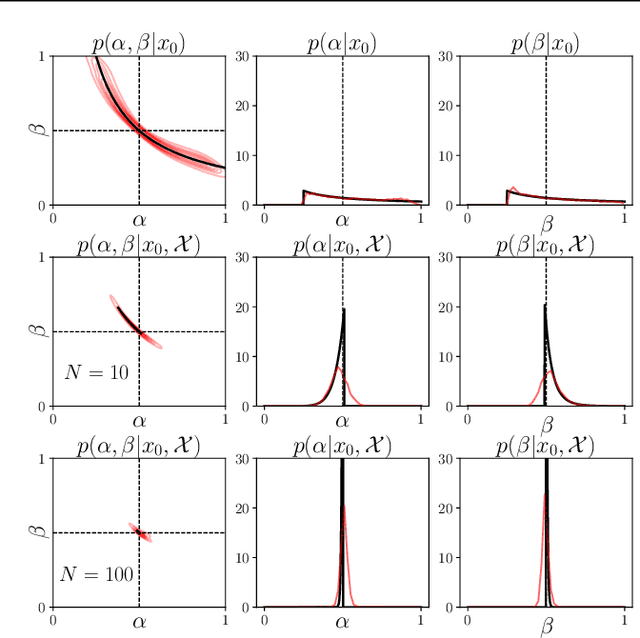
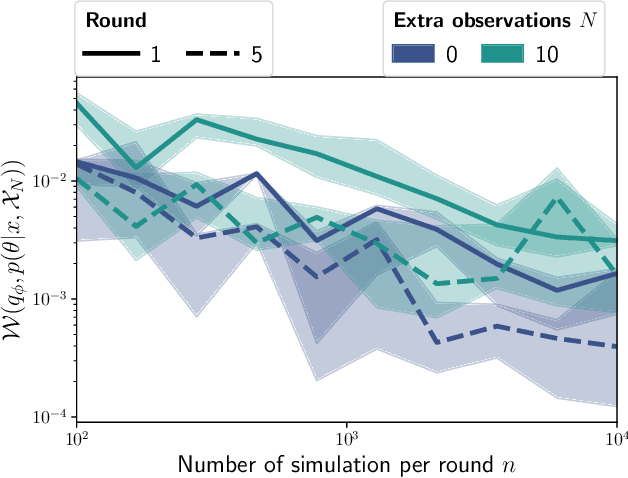
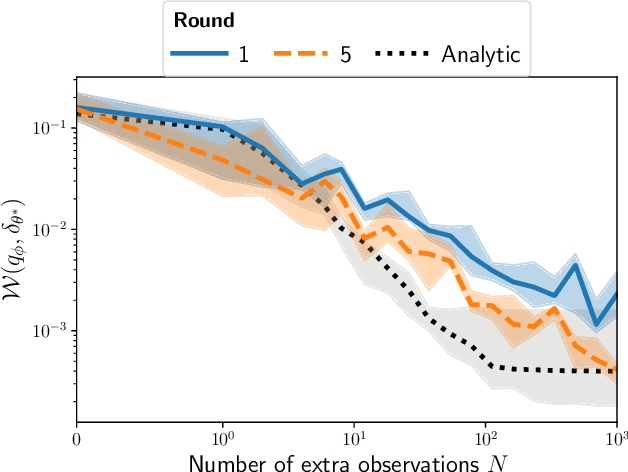
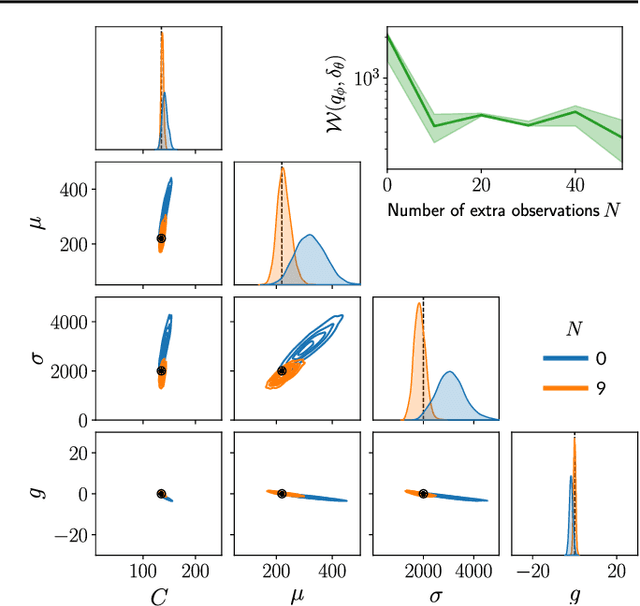
Abstract:Inferring the parameters of a stochastic model based on experimental observations is central to the scientific method. A particularly challenging setting is when the model is strongly indeterminate, i.e., when distinct sets of parameters yield identical observations. This arises in many practical situations, such as when inferring the distance and power of a radio source (is the source close and weak or far and strong?) or when estimating the amplifier gain and underlying brain activity of an electrophysiological experiment. In this work, we present a method for cracking such indeterminacy by exploiting additional information conveyed by an auxiliary set of observations sharing global parameters. Our method extends recent developments in simulation-based inference(SBI) based on normalizing flows to Bayesian hierarchical models. We validate quantitatively our proposal on a motivating example amenable to analytical solutions, and then apply it to invert a well known non-linear model from computational neuroscience.
Learning summary features of time series for likelihood free inference
Dec 04, 2020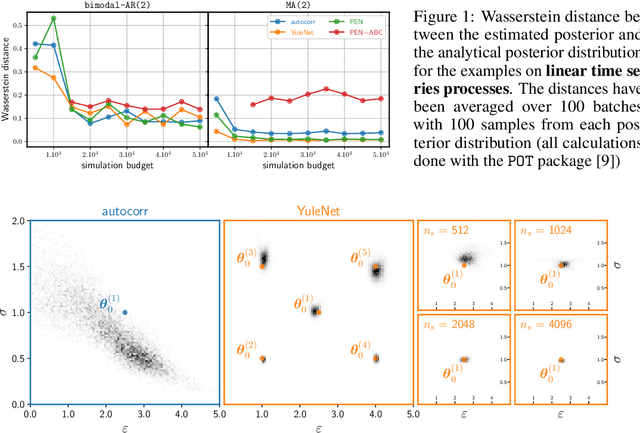
Abstract:There has been an increasing interest from the scientific community in using likelihood-free inference (LFI) to determine which parameters of a given simulator model could best describe a set of experimental data. Despite exciting recent results and a wide range of possible applications, an important bottleneck of LFI when applied to time series data is the necessity of defining a set of summary features, often hand-tailored based on domain knowledge. In this work, we present a data-driven strategy for automatically learning summary features from univariate time series and apply it to signals generated from autoregressive-moving-average (ARMA) models and the Van der Pol Oscillator. Our results indicate that learning summary features from data can compete and even outperform LFI methods based on hand-crafted values such as autocorrelation coefficients even in the linear case.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge