Paweł Włodarczyk-Pruszyński
RetroGNN: Approximating Retrosynthesis by Graph Neural Networks for De Novo Drug Design
Nov 25, 2020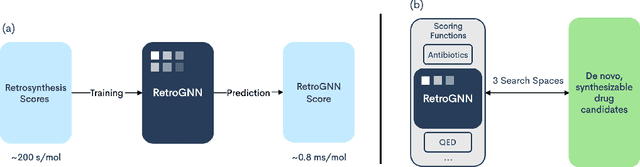
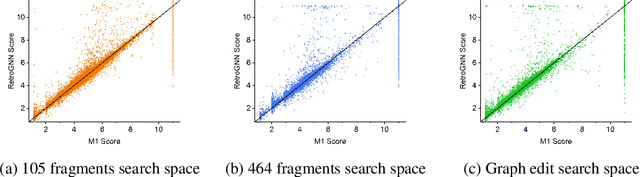
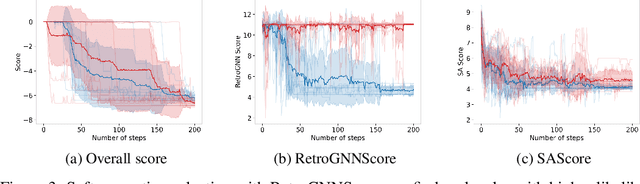
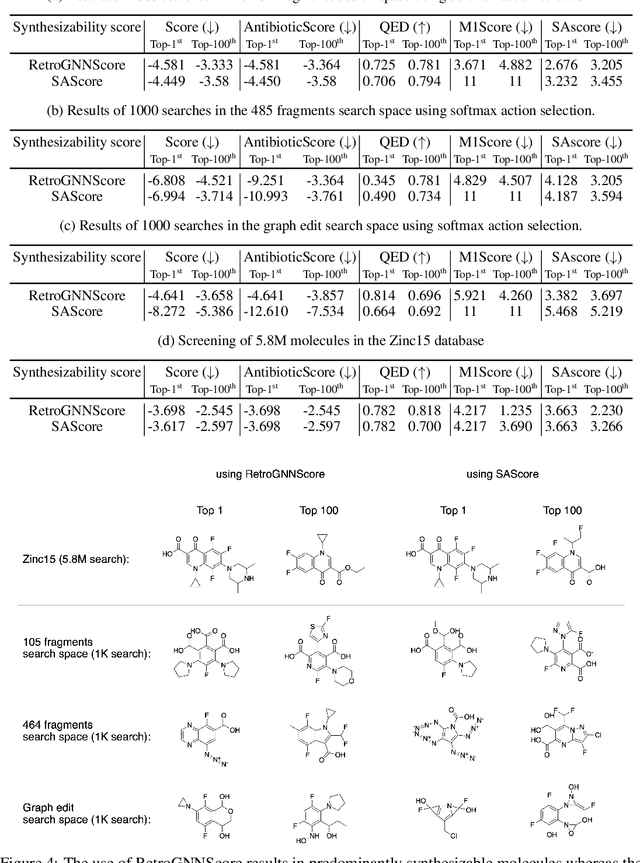
Abstract:De novo molecule generation often results in chemically unfeasible molecules. A natural idea to mitigate this problem is to bias the search process towards more easily synthesizable molecules using a proxy for synthetic accessibility. However, using currently available proxies still results in highly unrealistic compounds. We investigate the feasibility of training deep graph neural networks to approximate the outputs of a retrosynthesis planning software, and their use to bias the search process. We evaluate our method on a benchmark involving searching for drug-like molecules with antibiotic properties. Compared to enumerating over five million existing molecules from the ZINC database, our approach finds molecules predicted to be more likely to be antibiotics while maintaining good drug-like properties and being easily synthesizable. Importantly, our deep neural network can successfully filter out hard to synthesize molecules while achieving a $10^5$ times speed-up over using the retrosynthesis planning software.
Molecule Edit Graph Attention Network: Modeling Chemical Reactions as Sequences of Graph Edits
Jun 27, 2020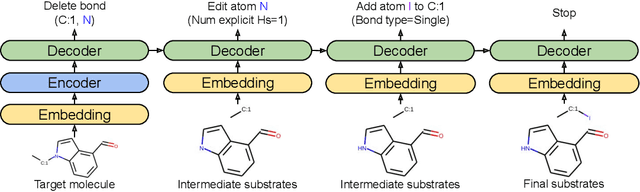
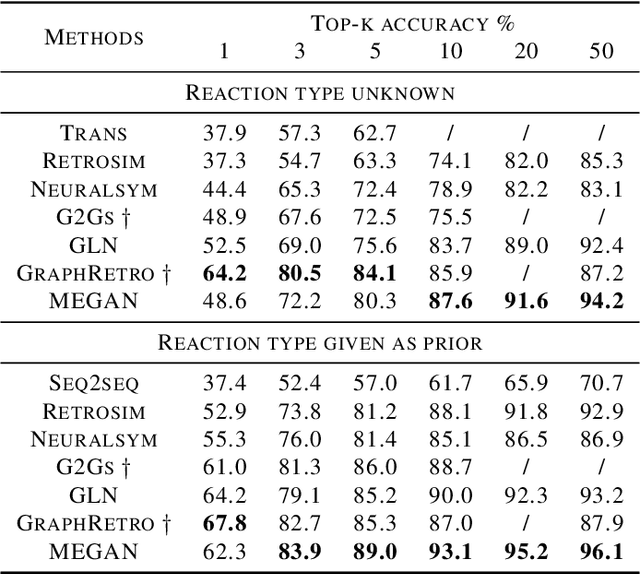
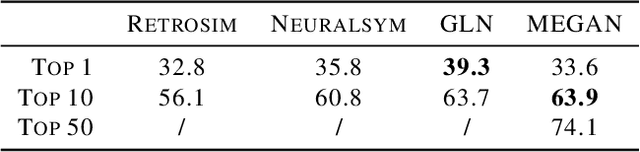
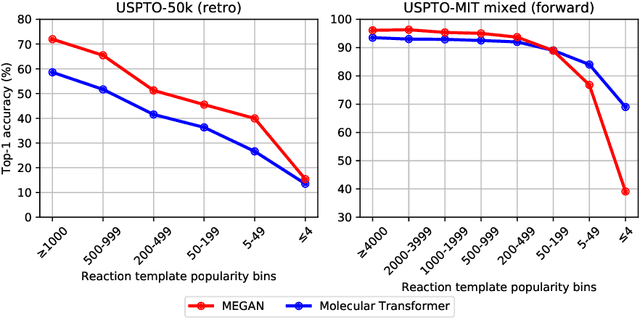
Abstract:One of the key challenges in automated synthesis planning is to generate diverse and reliable chemical reactions. Many reactions can be naturally represented using graph transformation rules referred broadly to as reaction templates. Using reaction templates enables accurate and interpretable predictions but can suffer from limited coverage of the reaction space. On the other hand, template-free methods can increase the coverage but can be prone to making trivial mistakes and are challenging to interpret. A promising idea for constructing more interpretable template-free models is to model a reaction as a sequence of graph edits of the substrates. We extend this idea to retrosynthesis and scale it up to large datasets. We propose Molecule Edit Graph Attention Network (MEGAN), a template-free neural model that encodes reaction as a sequence of graph edits. We achieve competitive performance on both retrosynthesis and forward synthesis and in particular state-of-the-art top-k accuracy for larger K values. Crucially, the latter shows excellent coverage of the reaction space of our model. In summary, MEGAN brings together the strong elements of template-free and template-based models and can be applied to both retro and forward synthesis tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge