Paulo Félix
Bayesian Nonparametric Dynamical Clustering of Time Series
Oct 08, 2025



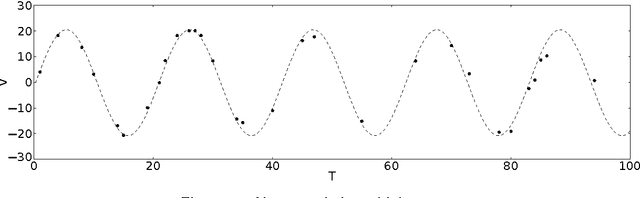
Abstract:We present a method that models the evolution of an unbounded number of time series clusters by switching among an unknown number of regimes with linear dynamics. We develop a Bayesian non-parametric approach using a hierarchical Dirichlet process as a prior on the parameters of a Switching Linear Dynamical System and a Gaussian process prior to model the statistical variations in amplitude and temporal alignment within each cluster. By modeling the evolution of time series patterns, the method avoids unnecessary proliferation of clusters in a principled manner. We perform inference by formulating a variational lower bound for off-line and on-line scenarios, enabling efficient learning through optimization. We illustrate the versatility and effectiveness of the approach through several case studies of electrocardiogram analysis using publicly available databases.
Hypothesis testing for matched pairs with missing data by maximum mean discrepancy: An application to continuous glucose monitoring
Jun 03, 2022



Abstract:A frequent problem in statistical science is how to properly handle missing data in matched paired observations. There is a large body of literature coping with the univariate case. Yet, the ongoing technological progress in measuring biological systems raises the need for addressing more complex data, e.g., graphs, strings and probability distributions, among others. In order to fill this gap, this paper proposes new estimators of the maximum mean discrepancy (MMD) to handle complex matched pairs with missing data. These estimators can detect differences in data distributions under different missingness mechanisms. The validity of this approach is proven and further studied in an extensive simulation study, and results of statistical consistency are provided. Data from continuous glucose monitoring in a longitudinal population-based diabetes study are used to illustrate the application of this approach. By employing the new distributional representations together with cluster analysis, new clinical criteria on how glucose changes vary at the distributional level over five years can be explored.
Glucose values prediction five years ahead with a new framework of missing responses in reproducing kernel Hilbert spaces, and the use of continuous glucose monitoring technology
Dec 14, 2020



Abstract:AEGIS study possesses unique information on longitudinal changes in circulating glucose through continuous glucose monitoring technology (CGM). However, as usual in longitudinal medical studies, there is a significant amount of missing data in the outcome variables. For example, 40 percent of glycosylated hemoglobin (A1C) biomarker data are missing five years ahead. With the purpose to reduce the impact of this issue, this article proposes a new data analysis framework based on learning in reproducing kernel Hilbert spaces (RKHS) with missing responses that allows to capture non-linear relations between variable studies in different supervised modeling tasks. First, we extend the Hilbert-Schmidt dependence measure to test statistical independence in this context introducing a new bootstrap procedure, for which we prove consistency. Next, we adapt or use existing models of variable selection, regression, and conformal inference to obtain new clinical findings about glucose changes five years ahead with the AEGIS data. The most relevant findings are summarized below: i) We identify new factors associated with long-term glucose evolution; ii) We show the clinical sensibility of CGM data to detect changes in glucose metabolism; iii) We can improve clinical interventions based on our algorithms' expected glucose changes according to patients' baseline characteristics.
On the adoption of abductive reasoning for time series interpretation
Jun 25, 2018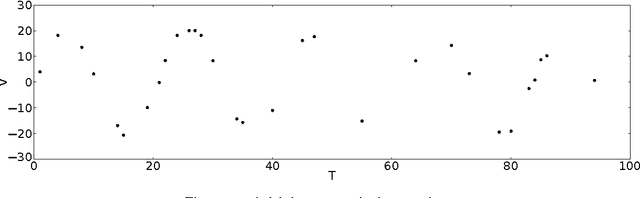
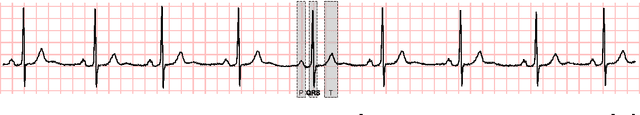
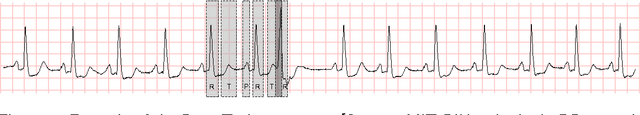
Abstract:Time series interpretation aims to provide an explanation of what is observed in terms of its underlying processes. The present work is based on the assumption that the common classification-based approaches to time series interpretation suffer from a set of inherent weaknesses, whose ultimate cause lies in the monotonic nature of the deductive reasoning paradigm. In this document we propose a new approach to this problem, based on the initial hypothesis that abductive reasoning properly accounts for the human ability to identify and characterize the patterns appearing in a time series. The result of this interpretation is a set of conjectures in the form of observations, organized into an abstraction hierarchy and explaining what has been observed. A knowledge-based framework and a set of algorithms for the interpretation task are provided, implementing a hypothesize-and-test cycle guided by an attentional mechanism. As a representative application domain, interpretation of the electrocardiogram allows us to highlight the strengths of the proposed approach in comparison with traditional classification-based approaches.
* 44 pages, 9 figures
Abductive reasoning as the basis to reproduce expert criteria in ECG Atrial Fibrillation identification
Feb 16, 2018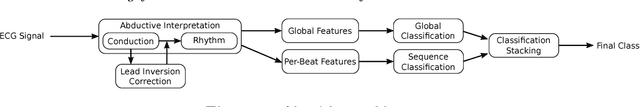
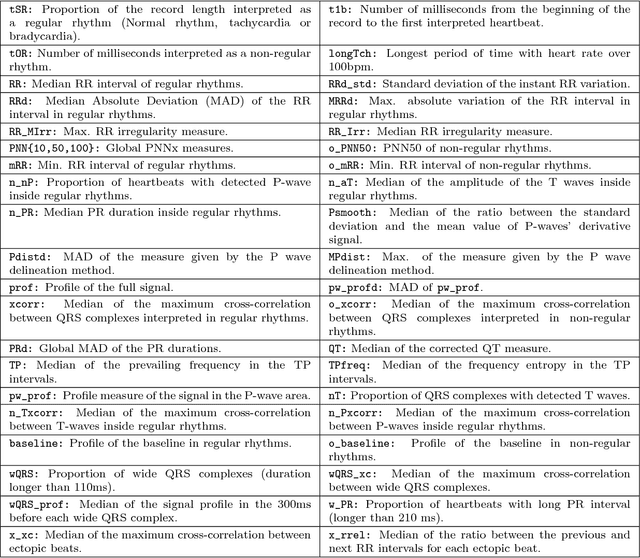
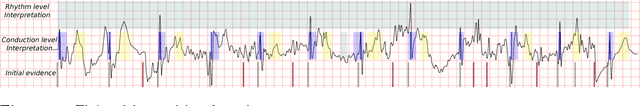
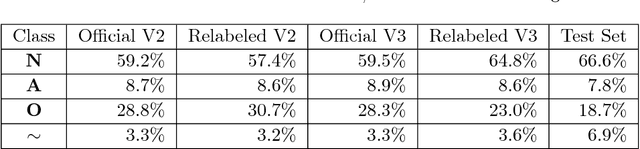
Abstract:Objective: This work aims at providing a new method for the automatic detection of atrial fibrillation, other arrhythmia and noise on short single lead ECG signals, emphasizing the importance of the interpretability of the classification results. Approach: A morphological and rhythm description of the cardiac behavior is obtained by a knowledge-based interpretation of the signal using the \textit{Construe} abductive framework. Then, a set of meaningful features are extracted for each individual heartbeat and as a summary of the full record. The feature distributions were used to elucidate the expert criteria underlying the labeling of the 2017 Physionet/CinC Challenge dataset, enabling a manual partial relabeling to improve the consistency of the classification rules. Finally, state-of-the-art machine learning methods are combined to provide an answer on the basis of the feature values. Main results: The proposal tied for the first place in the official stage of the Challenge, with a combined $F_1$ score of 0.83, and was even improved in the follow-up stage to 0.85 with a significant simplification of the model. Significance: This approach demonstrates the potential of \textit{Construe} to provide robust and valuable descriptions of temporal data even with significant amounts of noise and artifacts. Also, we discuss the importance of a consistent classification criteria in manually labeled training datasets, and the fundamental advantages of knowledge-based approaches to formalize and validate that criteria.
Arrhythmia Classification from the Abductive Interpretation of Short Single-Lead ECG Records
Nov 10, 2017
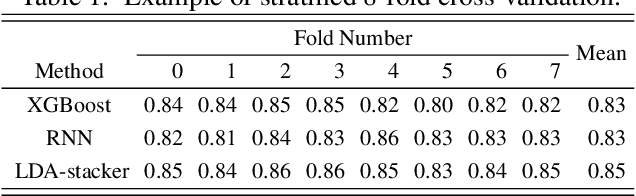
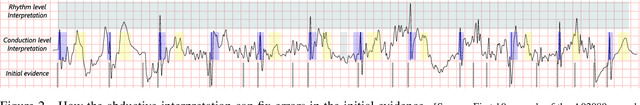
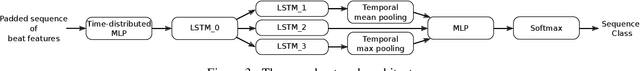
Abstract:In this work we propose a new method for the rhythm classification of short single-lead ECG records, using a set of high-level and clinically meaningful features provided by the abductive interpretation of the records. These features include morphological and rhythm-related features that are used to build two classifiers: one that evaluates the record globally, using aggregated values for each feature; and another one that evaluates the record as a sequence, using a Recurrent Neural Network fed with the individual features for each detected heartbeat. The two classifiers are finally combined using the stacking technique, providing an answer by means of four target classes: Normal sinus rhythm, Atrial fibrillation, Other anomaly, and Noisy. The approach has been validated against the 2017 Physionet/CinC Challenge dataset, obtaining a final score of 0.83 and ranking first in the competition.
Non-parametric Estimation of Stochastic Differential Equations with Sparse Gaussian Processes
Jul 10, 2017



Abstract:The application of Stochastic Differential Equations (SDEs) to the analysis of temporal data has attracted increasing attention, due to their ability to describe complex dynamics with physically interpretable equations. In this paper, we introduce a non-parametric method for estimating the drift and diffusion terms of SDEs from a densely observed discrete time series. The use of Gaussian processes as priors permits working directly in a function-space view and thus the inference takes place directly in this space. To cope with the computational complexity that requires the use of Gaussian processes, a sparse Gaussian process approximation is provided. This approximation permits the efficient computation of predictions for the drift and diffusion terms by using a distribution over a small subset of pseudo-samples. The proposed method has been validated using both simulated data and real data from economy and paleoclimatology. The application of the method to real data demonstrates its ability to capture the behaviour of complex systems.
Using temporal abduction for biosignal interpretation: A case study on QRS detection
Feb 05, 2015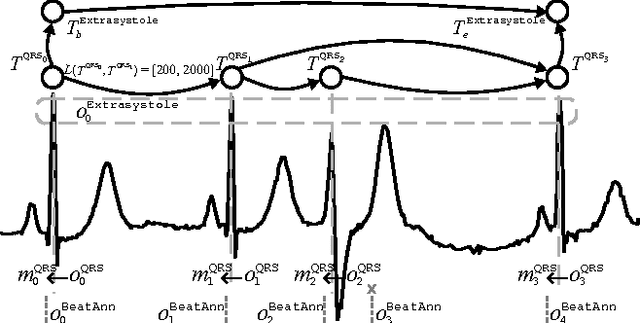
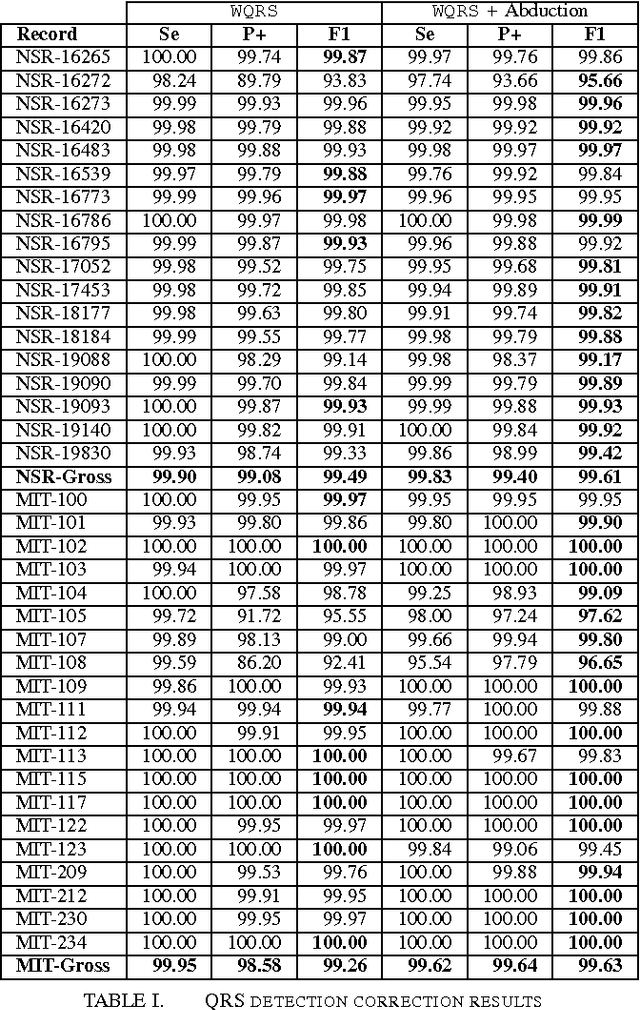
Abstract:In this work, we propose an abductive framework for biosignal interpretation, based on the concept of Temporal Abstraction Patterns. A temporal abstraction pattern defines an abstraction relation between an observation hypothesis and a set of observations constituting its evidence support. New observations are generated abductively from any subset of the evidence of a pattern, building an abstraction hierarchy of observations in which higher levels contain those observations with greater interpretative value of the physiological processes underlying a given signal. Non-monotonic reasoning techniques have been applied to this model in order to find the best interpretation of a set of initial observations, permitting even to correct these observations by removing, adding or modifying them in order to make them consistent with the available domain knowledge. Some preliminary experiments have been conducted to apply this framework to a well known and bounded problem: the QRS detection on ECG signals. The objective is not to provide a new better QRS detector, but to test the validity of an abductive paradigm. These experiments show that a knowledge base comprising just a few very simple rhythm abstraction patterns can enhance the results of a state of the art algorithm by significantly improving its detection F1-score, besides proving the ability of the abductive framework to correct both sensitivity and specificity failures.
A method for context-based adaptive QRS clustering in real-time
Oct 27, 2014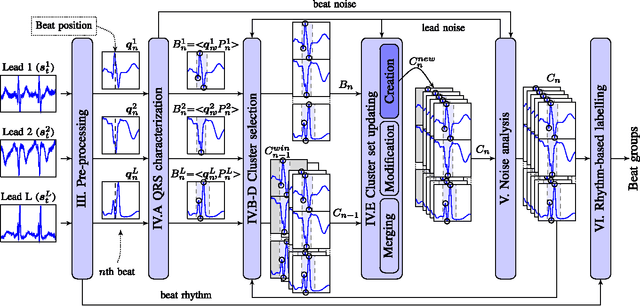

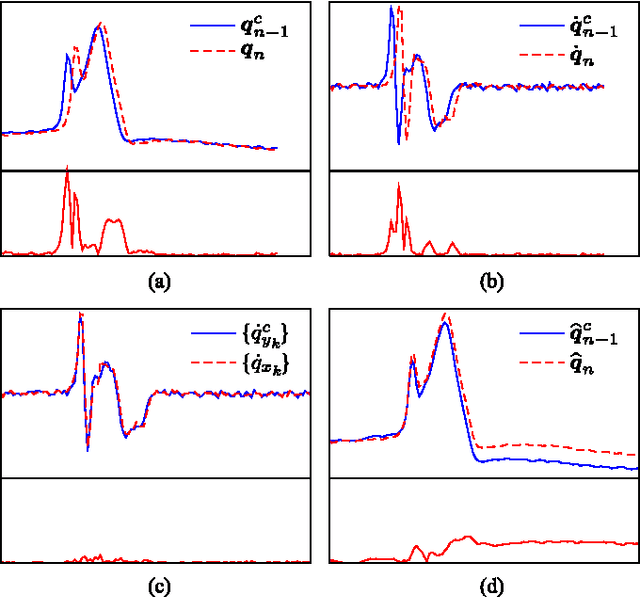

Abstract:Continuous follow-up of heart condition through long-term electrocardiogram monitoring is an invaluable tool for diagnosing some cardiac arrhythmias. In such context, providing tools for fast locating alterations of normal conduction patterns is mandatory and still remains an open issue. This work presents a real-time method for adaptive clustering QRS complexes from multilead ECG signals that provides the set of QRS morphologies that appear during an ECG recording. The method processes the QRS complexes sequentially, grouping them into a dynamic set of clusters based on the information content of the temporal context. The clusters are represented by templates which evolve over time and adapt to the QRS morphology changes. Rules to create, merge and remove clusters are defined along with techniques for noise detection in order to avoid their proliferation. To cope with beat misalignment, Derivative Dynamic Time Warping is used. The proposed method has been validated against the MIT-BIH Arrhythmia Database and the AHA ECG Database showing a global purity of 98.56% and 99.56%, respectively. Results show that our proposal not only provides better results than previous offline solutions but also fulfills real-time requirements.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge