Jesús Presedo
Bayesian Nonparametric Dynamical Clustering of Time Series
Oct 08, 2025



Abstract:We present a method that models the evolution of an unbounded number of time series clusters by switching among an unknown number of regimes with linear dynamics. We develop a Bayesian non-parametric approach using a hierarchical Dirichlet process as a prior on the parameters of a Switching Linear Dynamical System and a Gaussian process prior to model the statistical variations in amplitude and temporal alignment within each cluster. By modeling the evolution of time series patterns, the method avoids unnecessary proliferation of clusters in a principled manner. We perform inference by formulating a variational lower bound for off-line and on-line scenarios, enabling efficient learning through optimization. We illustrate the versatility and effectiveness of the approach through several case studies of electrocardiogram analysis using publicly available databases.
Non-parametric Estimation of Stochastic Differential Equations with Sparse Gaussian Processes
Jul 10, 2017



Abstract:The application of Stochastic Differential Equations (SDEs) to the analysis of temporal data has attracted increasing attention, due to their ability to describe complex dynamics with physically interpretable equations. In this paper, we introduce a non-parametric method for estimating the drift and diffusion terms of SDEs from a densely observed discrete time series. The use of Gaussian processes as priors permits working directly in a function-space view and thus the inference takes place directly in this space. To cope with the computational complexity that requires the use of Gaussian processes, a sparse Gaussian process approximation is provided. This approximation permits the efficient computation of predictions for the drift and diffusion terms by using a distribution over a small subset of pseudo-samples. The proposed method has been validated using both simulated data and real data from economy and paleoclimatology. The application of the method to real data demonstrates its ability to capture the behaviour of complex systems.
Using temporal abduction for biosignal interpretation: A case study on QRS detection
Feb 05, 2015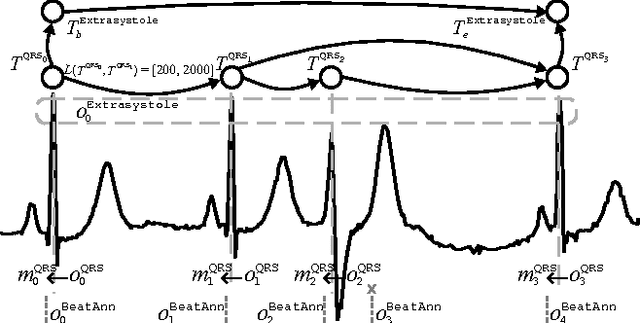
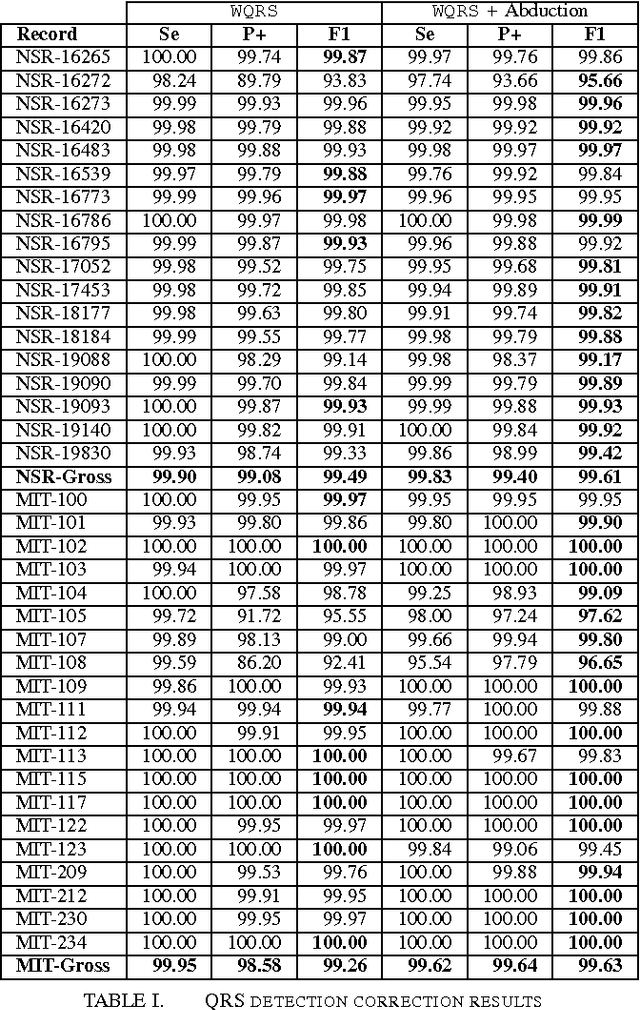
Abstract:In this work, we propose an abductive framework for biosignal interpretation, based on the concept of Temporal Abstraction Patterns. A temporal abstraction pattern defines an abstraction relation between an observation hypothesis and a set of observations constituting its evidence support. New observations are generated abductively from any subset of the evidence of a pattern, building an abstraction hierarchy of observations in which higher levels contain those observations with greater interpretative value of the physiological processes underlying a given signal. Non-monotonic reasoning techniques have been applied to this model in order to find the best interpretation of a set of initial observations, permitting even to correct these observations by removing, adding or modifying them in order to make them consistent with the available domain knowledge. Some preliminary experiments have been conducted to apply this framework to a well known and bounded problem: the QRS detection on ECG signals. The objective is not to provide a new better QRS detector, but to test the validity of an abductive paradigm. These experiments show that a knowledge base comprising just a few very simple rhythm abstraction patterns can enhance the results of a state of the art algorithm by significantly improving its detection F1-score, besides proving the ability of the abductive framework to correct both sensitivity and specificity failures.
A method for context-based adaptive QRS clustering in real-time
Oct 27, 2014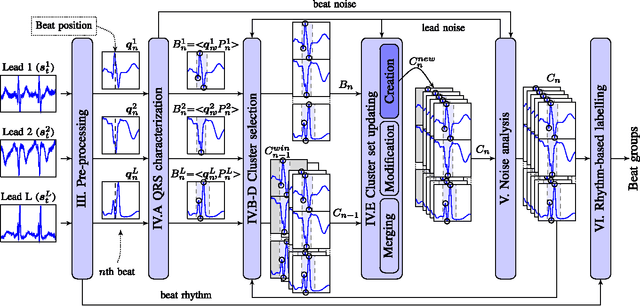

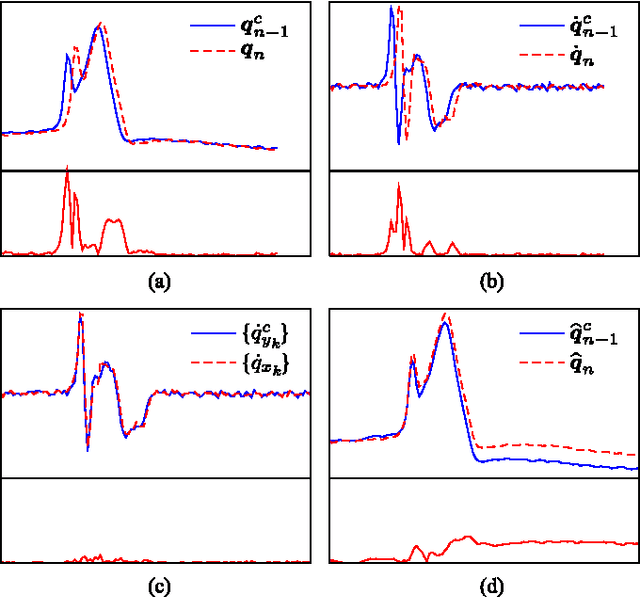

Abstract:Continuous follow-up of heart condition through long-term electrocardiogram monitoring is an invaluable tool for diagnosing some cardiac arrhythmias. In such context, providing tools for fast locating alterations of normal conduction patterns is mandatory and still remains an open issue. This work presents a real-time method for adaptive clustering QRS complexes from multilead ECG signals that provides the set of QRS morphologies that appear during an ECG recording. The method processes the QRS complexes sequentially, grouping them into a dynamic set of clusters based on the information content of the temporal context. The clusters are represented by templates which evolve over time and adapt to the QRS morphology changes. Rules to create, merge and remove clusters are defined along with techniques for noise detection in order to avoid their proliferation. To cope with beat misalignment, Derivative Dynamic Time Warping is used. The proposed method has been validated against the MIT-BIH Arrhythmia Database and the AHA ECG Database showing a global purity of 98.56% and 99.56%, respectively. Results show that our proposal not only provides better results than previous offline solutions but also fulfills real-time requirements.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge