Padma Kaul
Exploring Best Practices for ECG Signal Processing in Machine Learning
Nov 02, 2023Abstract:In this work we search for best practices in pre-processing of Electrocardiogram (ECG) signals in order to train better classifiers for the diagnosis of heart conditions. State of the art machine learning algorithms have achieved remarkable results in classification of some heart conditions using ECG data, yet there appears to be no consensus on pre-processing best practices. Is this lack of consensus due to different conditions and architectures requiring different processing steps for optimal performance? Is it possible that state of the art deep-learning models have rendered pre-processing unnecessary? In this work we apply down-sampling, normalization, and filtering functions to 3 different multi-label ECG datasets and measure their effects on 3 different high-performing time-series classifiers. We find that sampling rates as low as 50Hz can yield comparable results to the commonly used 500Hz. This is significant as smaller sampling rates will result in smaller datasets and models, which require less time and resources to train. Additionally, despite their common usage, we found min-max normalization to be slightly detrimental overall, and band-passing to make no measurable difference. We found the blind approach to pre-processing of ECGs for multi-label classification to be ineffective, with the exception of sample rate reduction which reliably reduces computational resources, but does not increase accuracy.
Improving ECG-based COVID-19 diagnosis and mortality predictions using pre-pandemic medical records at population-scale
Nov 14, 2022



Abstract:Pandemic outbreaks such as COVID-19 occur unexpectedly, and need immediate action due to their potential devastating consequences on global health. Point-of-care routine assessments such as electrocardiogram (ECG), can be used to develop prediction models for identifying individuals at risk. However, there is often too little clinically-annotated medical data, especially in early phases of a pandemic, to develop accurate prediction models. In such situations, historical pre-pandemic health records can be utilized to estimate a preliminary model, which can then be fine-tuned based on limited available pandemic data. This study shows this approach -- pre-train deep learning models with pre-pandemic data -- can work effectively, by demonstrating substantial performance improvement over three different COVID-19 related diagnostic and prognostic prediction tasks. Similar transfer learning strategies can be useful for developing timely artificial intelligence solutions in future pandemic outbreaks.
ECG for high-throughput screening of multiple diseases: Proof-of-concept using multi-diagnosis deep learning from population-based datasets
Oct 06, 2022
Abstract:Electrocardiogram (ECG) abnormalities are linked to cardiovascular diseases, but may also occur in other non-cardiovascular conditions such as mental, neurological, metabolic and infectious conditions. However, most of the recent success of deep learning (DL) based diagnostic predictions in selected patient cohorts have been limited to a small set of cardiac diseases. In this study, we use a population-based dataset of >250,000 patients with >1000 medical conditions and >2 million ECGs to identify a wide range of diseases that could be accurately diagnosed from the patient's first in-hospital ECG. Our DL models uncovered 128 diseases and 68 disease categories with strong discriminative performance.
Multilabel 12-Lead Electrocardiogram Classification Using Gradient Boosting Tree Ensemble
Oct 21, 2020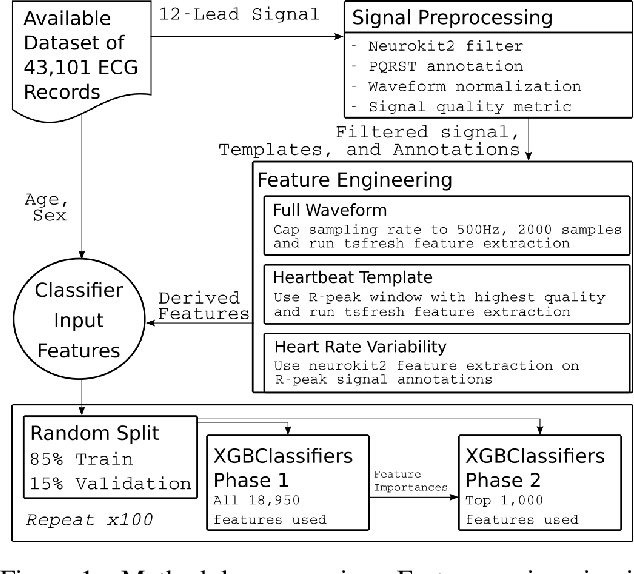
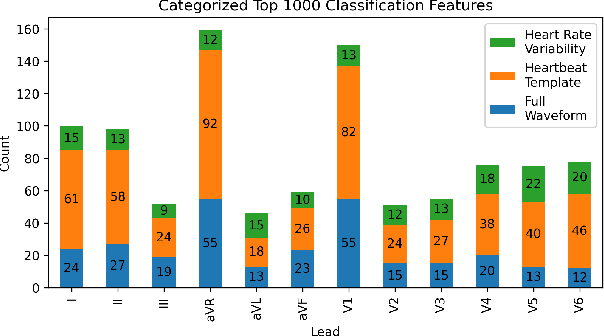
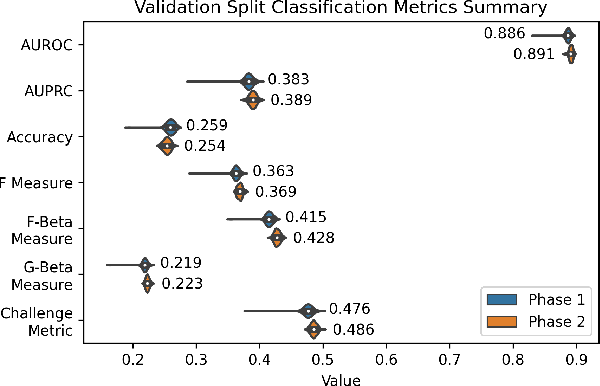
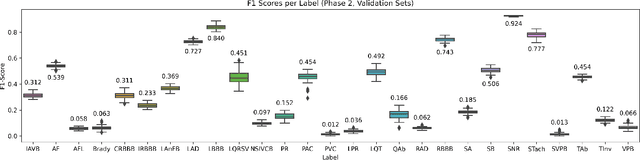
Abstract:The 12-lead electrocardiogram (ECG) is a commonly used tool for detecting cardiac abnormalities such as atrial fibrillation, blocks, and irregular complexes. For the PhysioNet/CinC 2020 Challenge, we built an algorithm using gradient boosted tree ensembles fitted on morphology and signal processing features to classify ECG diagnosis. For each lead, we derive features from heart rate variability, PQRST template shape, and the full signal waveform. We join the features of all 12 leads to fit an ensemble of gradient boosting decision trees to predict probabilities of ECG instances belonging to each class. We train a phase one set of feature importance determining models to isolate the top 1,000 most important features to use in our phase two diagnosis prediction models. We use repeated random sub-sampling by splitting our dataset of 43,101 records into 100 independent runs of 85:15 training/validation splits for our internal evaluation results. Our methodology generates us an official phase validation set score of 0.476 and test set score of -0.080 under the team name, CVC, placing us 36 out of 41 in the rankings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge