P. A. N. Bosman
Temporal True and Surrogate Fitness Landscape Analysis for Expensive Bi-Objective Optimisation
Apr 09, 2024Abstract:Many real-world problems have expensive-to-compute fitness functions and are multi-objective in nature. Surrogate-assisted evolutionary algorithms are often used to tackle such problems. Despite this, literature about analysing the fitness landscapes induced by surrogate models is limited, and even non-existent for multi-objective problems. This study addresses this critical gap by comparing landscapes of the true fitness function with those of surrogate models for multi-objective functions. Moreover, it does so temporally by examining landscape features at different points in time during optimisation, in the vicinity of the population at that point in time. We consider the BBOB bi-objective benchmark functions in our experiments. The results of the fitness landscape analysis reveals significant differences between true and surrogate features at different time points during optimisation. Despite these differences, the true and surrogate landscape features still show high correlations between each other. Furthermore, this study identifies which landscape features are related to search and demonstrates that both surrogate and true landscape features are capable of predicting algorithm performance. These findings indicate that temporal analysis of the landscape features may help to facilitate the design of surrogate switching approaches to improve performance in multi-objective optimisation.
Function Class Learning with Genetic Programming: Towards Explainable Meta Learning for Tumor Growth Functionals
Feb 19, 2024

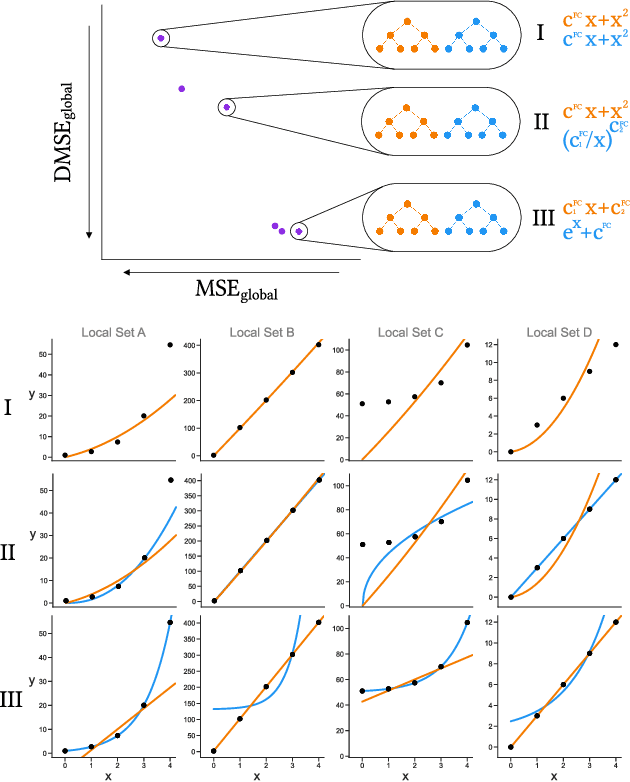
Abstract:Paragangliomas are rare, primarily slow-growing tumors for which the underlying growth pattern is unknown. Therefore, determining the best care for a patient is hard. Currently, if no significant tumor growth is observed, treatment is often delayed, as treatment itself is not without risk. However, by doing so, the risk of (irreversible) adverse effects due to tumor growth may increase. Being able to predict the growth accurately could assist in determining whether a patient will need treatment during their lifetime and, if so, the timing of this treatment. The aim of this work is to learn the general underlying growth pattern of paragangliomas from multiple tumor growth data sets, in which each data set contains a tumor's volume over time. To do so, we propose a novel approach based on genetic programming to learn a function class, i.e., a parameterized function that can be fit anew for each tumor. We do so in a unique, multi-modal, multi-objective fashion to find multiple potentially interesting function classes in a single run. We evaluate our approach on a synthetic and a real-world data set. By analyzing the resulting function classes, we can effectively explain the general patterns in the data.
Multi-modal multi-objective model-based genetic programming to find multiple diverse high-quality models
Mar 24, 2022

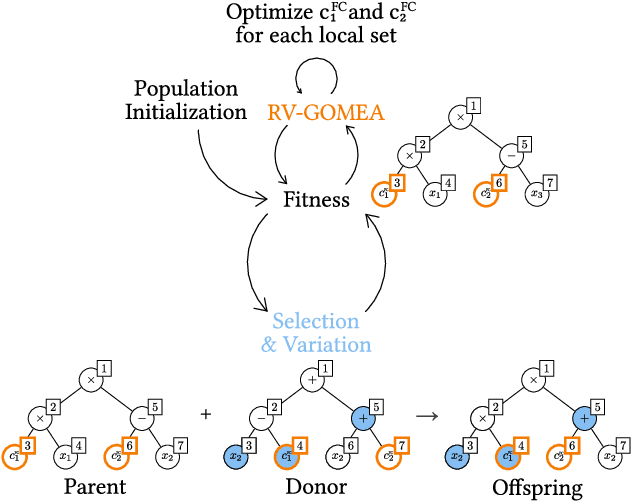
Abstract:Explainable artificial intelligence (XAI) is an important and rapidly expanding research topic. The goal of XAI is to gain trust in a machine learning (ML) model through clear insights into how the model arrives at its predictions. Genetic programming (GP) is often cited as being uniquely well-suited to contribute to XAI because of its capacity to learn (small) symbolic models that have the potential to be interpreted. Nevertheless, like many ML algorithms, GP typically results in a single best model. However, in practice, the best model in terms of training error may well not be the most suitable one as judged by a domain expert for various reasons, including overfitting, multiple different models existing that have similar accuracy, and unwanted errors on particular data points due to typical accuracy measures like mean squared error. Hence, to increase chances that domain experts deem a resulting model plausible, it becomes important to be able to explicitly search for multiple, diverse, high-quality models that trade-off different meanings of accuracy. In this paper, we achieve exactly this with a novel multi-modal multi-tree multi-objective GP approach that extends a modern model-based GP algorithm known as GP-GOMEA that is already effective at searching for small expressions.
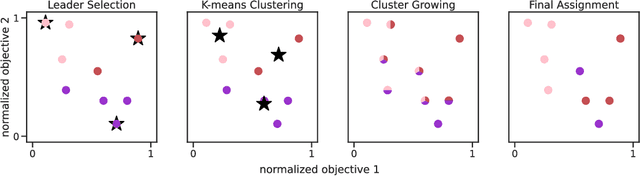
Real-valued Evolutionary Multi-modal Multi-objective Optimization by Hill-Valley Clustering
Oct 28, 2020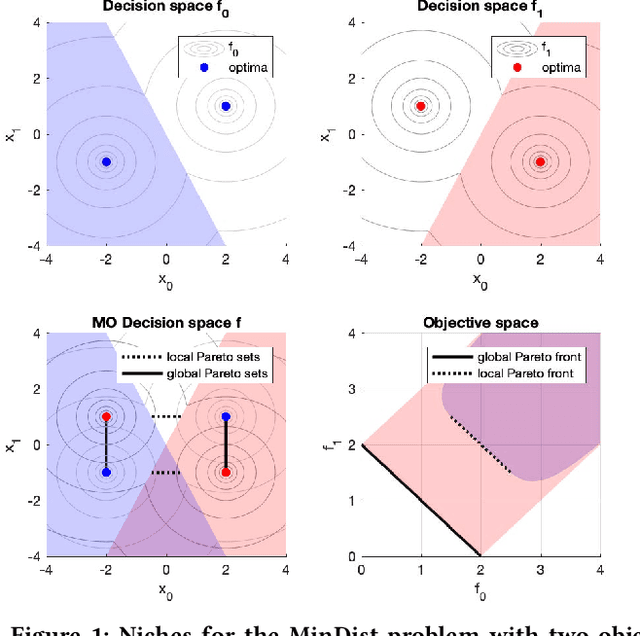
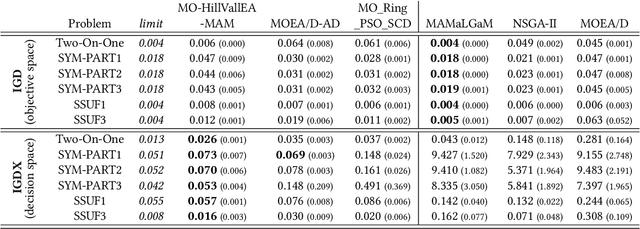

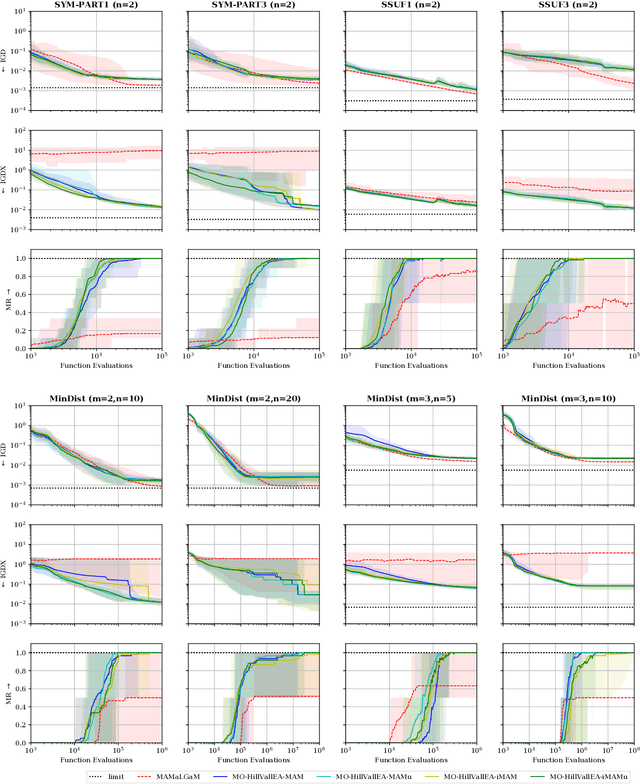
Abstract:In model-based evolutionary algorithms (EAs), the underlying search distribution is adapted to the problem at hand, for example based on dependencies between decision variables. Hill-valley clustering is an adaptive niching method in which a set of solutions is clustered such that each cluster corresponds to a single mode in the fitness landscape. This can be used to adapt the search distribution of an EA to the number of modes, exploring each mode separately. Especially in a black-box setting, where the number of modes is a priori unknown, an adaptive approach is essential for good performance. In this work, we introduce multi-objective hill-valley clustering and combine it with MAMaLGaM, a multi-objective EA, into the multi-objective hill-valley EA (MO-HillVallEA). We empirically show that MO-HillVallEA outperforms MAMaLGaM and other well-known multi-objective optimization algorithms on a set of benchmark functions. Furthermore, and perhaps most important, we show that MO-HillVallEA is capable of obtaining and maintaining multiple approximation sets simultaneously over time.
Ensuring smoothly navigable approximation sets by Bezier curve parameterizations in evolutionary bi-objective optimization -- applied to brachytherapy treatment planning for prostate cancer
Jun 11, 2020


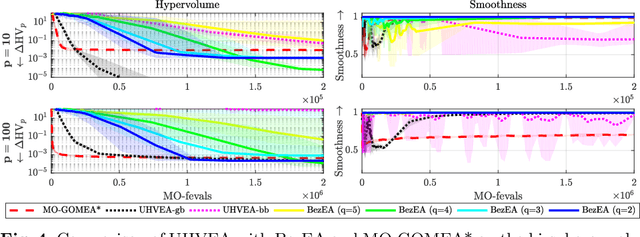
Abstract:The aim of bi-objective optimization is to obtain an approximation set of (near) Pareto optimal solutions. A decision maker then navigates this set to select a final desired solution, often using a visualization of the approximation front. The front provides a navigational ordering of solutions to traverse, but this ordering does not necessarily map to a smooth trajectory through decision space. This forces the decision maker to inspect the decision variables of each solution individually, potentially making navigation of the approximation set unintuitive. In this work, we aim to improve approximation set navigability by enforcing a form of smoothness or continuity between solutions in terms of their decision variables. Imposing smoothness as a restriction upon common domination-based multi-objective evolutionary algorithms is not straightforward. Therefore, we use the recently introduced uncrowded hypervolume (UHV) to reformulate the multi-objective optimization problem as a single-objective problem in which parameterized approximation sets are directly optimized. We study here the case of parameterizing approximation sets as smooth Bezier curves in decision space. We approach the resulting single-objective problem with the gene-pool optimal mixing evolutionary algorithm (GOMEA), and we call the resulting algorithm BezEA. We analyze the behavior of BezEA and compare it to optimization of the UHV with GOMEA as well as the domination-based multi-objective GOMEA. We show that high-quality approximation sets can be obtained with BezEA, sometimes even outperforming the domination- and UHV-based algorithms, while smoothness of the navigation trajectory through decision space is guaranteed.
Local Search is a Remarkably Strong Baseline for Neural Architecture Search
Apr 29, 2020

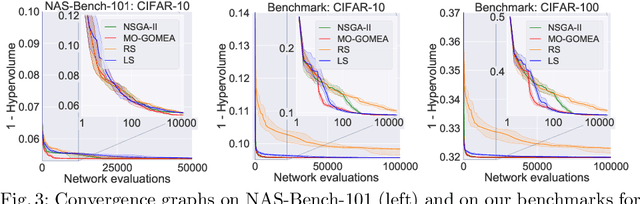
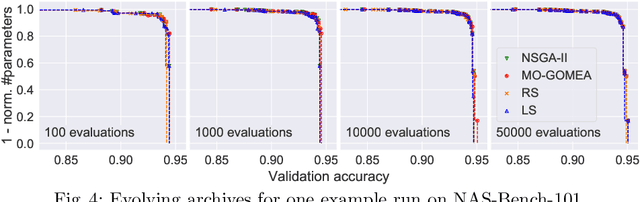
Abstract:Neural Architecture Search (NAS), i.e., the automation of neural network design, has gained much popularity in recent years with increasingly complex search algorithms being proposed. Yet, solid comparisons with simple baselines are often missing. At the same time, recent retrospective studies have found many new algorithms to be no better than random search (RS). In this work we consider, for the first time, a simple Local Search (LS) algorithm for NAS. We particularly consider a multi-objective NAS formulation, with network accuracy and network complexity as two objectives, as understanding the trade-off between these two objectives is arguably the most interesting aspect of NAS. The proposed LS algorithm is compared with RS and two evolutionary algorithms (EAs), as these are often heralded as being ideal for multi-objective optimization. To promote reproducibility, we create and release two benchmark datasets containing 200K saved network evaluations for two established image classification tasks, CIFAR-10 and CIFAR-100. Our benchmarks are designed to be complementary to existing benchmarks, especially in that they are better suited for multi-objective search. We additionally consider a version of the problem with a much larger architecture space. While we find and show that the considered algorithms explore the search space in fundamentally different ways, we also find that LS substantially outperforms RS and even performs nearly as good as state-of-the-art EAs. We believe that this provides strong evidence that LS is truly a competitive baseline for NAS against which new NAS algorithms should be benchmarked.
Uncrowded Hypervolume-based Multi-objective Optimization with Gene-pool Optimal Mixing
Apr 10, 2020
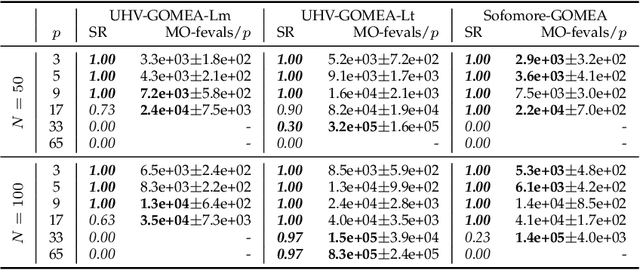
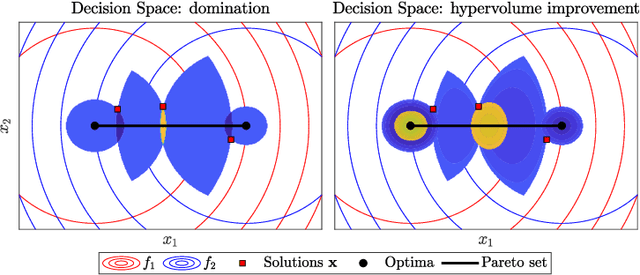
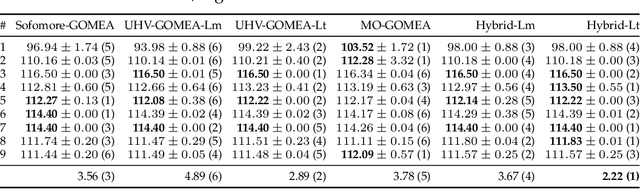
Abstract:Domination-based multi-objective (MO) evolutionary algorithms (EAs) are today arguably the most frequently used type of MOEA. These methods however stagnate when the majority of the population becomes non-dominated, preventing convergence to the Pareto set. Hypervolume-based MO optimization has shown promising results to overcome this. Direct use of the hypervolume however results in no selection pressure for dominated solutions. The recently introduced Sofomore framework overcomes this by solving multiple interleaved single-objective dynamic problems that iteratively improve a single approximation set, based on the uncrowded hypervolume improvement (UHVI). It thereby however loses many advantages of population-based MO optimization, such as handling multimodality. Here, we reformulate the UHVI as a quality measure for approximation sets, called the uncrowded hypervolume (UHV), which can be used to directly solve MO optimization problems with a single-objective optimizer. We use the state-of-the-art gene-pool optimal mixing evolutionary algorithm (GOMEA) that is capable of efficiently exploiting the intrinsically available grey-box properties of this problem. The resulting algorithm, UHV-GOMEA, is compared to Sofomore equipped with GOMEA, and the domination-based MO-GOMEA. In doing so, we investigate in which scenarios either domination-based or hypervolume-based methods are preferred. Finally, we construct a simple hybrid approach that combines MO-GOMEA with UHV-GOMEA and outperforms both.
Surrogate-free machine learning-based organ dose reconstruction for pediatric abdominal radiotherapy
Feb 17, 2020



Abstract:To study radiotherapy-related adverse effects, detailed dose information (3D distribution) is needed for accurate dose-effect modeling. For childhood cancer survivors who underwent radiotherapy in the pre-CT era, only 2D radiographs were acquired, thus 3D dose distributions must be reconstructed. State-of-the-art methods achieve this by using 3D surrogate anatomies. These can however lack personalization and lead to coarse reconstructions. We present and validate a surrogate-free dose reconstruction method based on Machine Learning (ML). Abdominal planning CTs (n=142) of recently-treated childhood cancer patients were gathered, their organs at risk were segmented, and 300 artificial Wilms' tumor plans were sampled automatically. Each artificial plan was automatically emulated on the 142 CTs, resulting in 42,600 3D dose distributions from which dose-volume metrics were derived. Anatomical features were extracted from digitally reconstructed radiographs simulated from the CTs to resemble historical radiographs. Further, patient and radiotherapy plan features typically available from historical treatment records were collected. An evolutionary ML algorithm was then used to link features to dose-volume metrics. Besides 5-fold cross validation, a further evaluation was done on an independent dataset of five CTs each associated with two clinical plans. Cross-validation resulted in mean absolute errors $\leq$0.6 Gy for organs completely inside or outside the field. For organs positioned at the edge of the field, mean absolute errors $\leq$1.7 Gy for $D_{mean}$, $\leq$2.9 Gy for $D_{2cc}$, and $\leq$13% for $V_{5Gy}$ and $V_{10Gy}$, were obtained, without systematic bias. Similar results were found for the independent dataset. To conclude, our novel organ dose reconstruction method is not only accurate, but also efficient, as the setup of a surrogate is no longer needed.
Benchmarking HillVallEA for the GECCO 2019 Competition on Multimodal Optimization
Jul 25, 2019
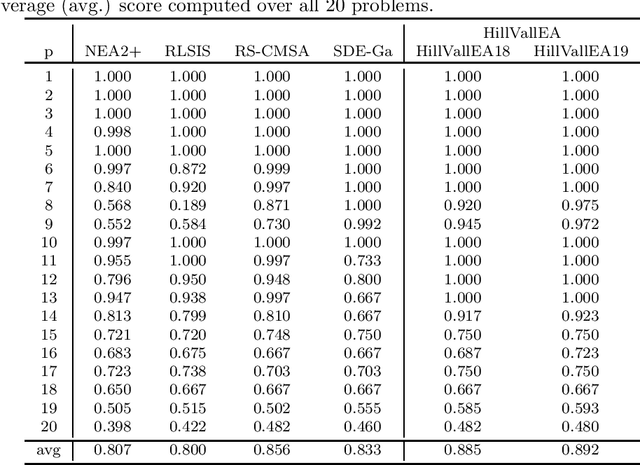
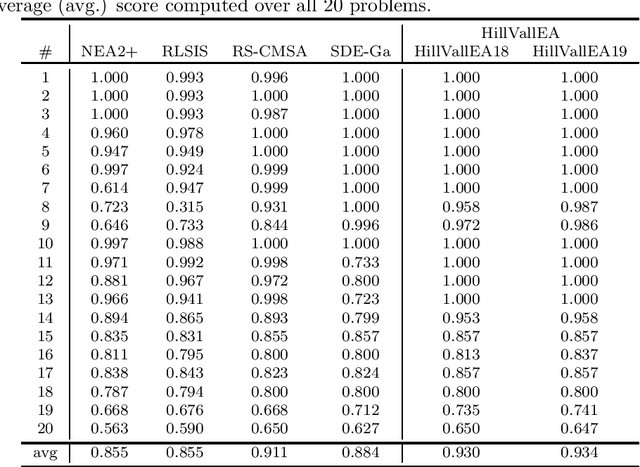
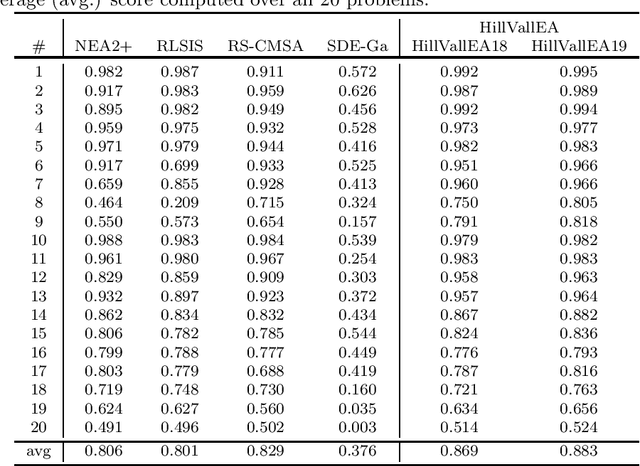
Abstract:This report presents benchmarking results of the Hill-Valley Evolutionary Algorithm version 2019 (HillVallEA19) on the CEC2013 niching benchmark suite under the restrictions of the GECCO 2019 niching competition on multimodal optimization. Performance is compared to algorithms that participated in previous editions of the niching competition.
Real-Valued Evolutionary Multi-Modal Optimization driven by Hill-Valley Clustering
Oct 16, 2018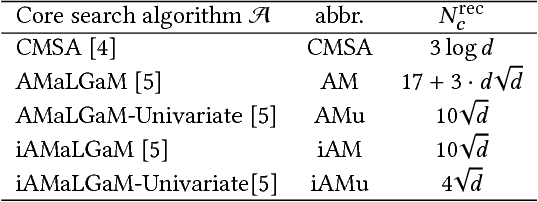
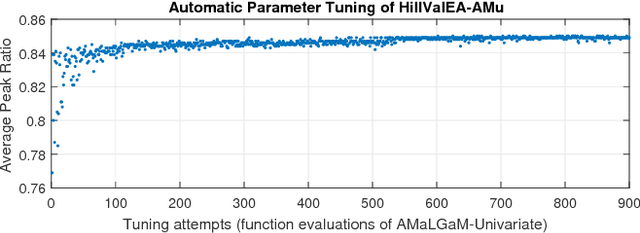
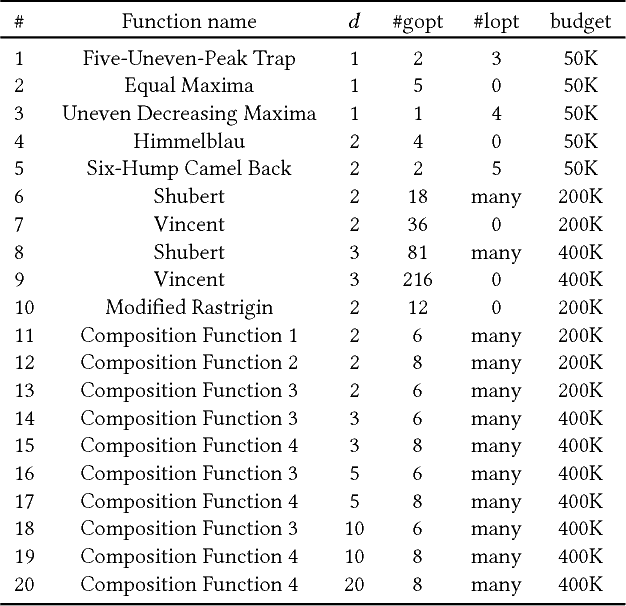
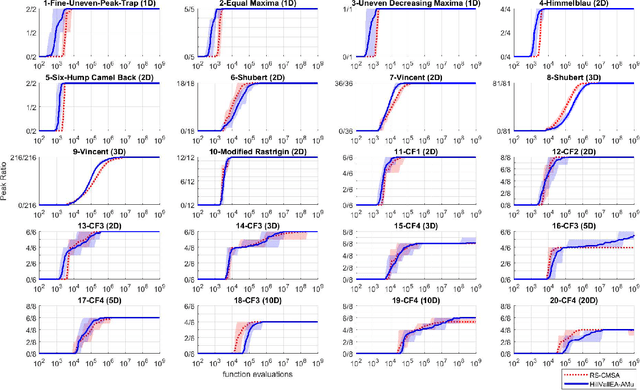
Abstract:Model-based evolutionary algorithms (EAs) adapt an underlying search model to features of the problem at hand, such as the linkage between problem variables. The performance of EAs often deteriorates as multiple modes in the fitness landscape are modelled with a unimodal search model. The number of modes is however often unknown a priori, especially in a black-box setting, which complicates adaptation of the search model. In this work, we focus on models that can adapt to the multi-modality of the fitness landscape. Specifically, we introduce Hill-Valley Clustering, a remarkably simple approach to adaptively cluster the search space in niches, such that a single mode resides in each niche. In each of the located niches, a core search algorithm is initialized to optimize that niche. Combined with an EA and a restart scheme, the resulting Hill-Valley EA (HillVallEA) is compared to current state-of-the-art niching methods on a standard benchmark suite for multi-modal optimization. Numerical results in terms of the detected number of global optima show that, in spite of its simplicity, HillVallEA is competitive within the limited budget of the benchmark suite, and shows superior performance in the long run.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge