M. Virgolin
Interpretable Symbolic Regression for Data Science: Analysis of the 2022 Competition
Apr 06, 2023



Abstract:Symbolic regression searches for analytic expressions that accurately describe studied phenomena. The main attraction of this approach is that it returns an interpretable model that can be insightful to users. Historically, the majority of algorithms for symbolic regression have been based on evolutionary algorithms. However, there has been a recent surge of new proposals that instead utilize approaches such as enumeration algorithms, mixed linear integer programming, neural networks, and Bayesian optimization. In order to assess how well these new approaches behave on a set of common challenges often faced in real-world data, we hosted a competition at the 2022 Genetic and Evolutionary Computation Conference consisting of different synthetic and real-world datasets which were blind to entrants. For the real-world track, we assessed interpretability in a realistic way by using a domain expert to judge the trustworthiness of candidate models.We present an in-depth analysis of the results obtained in this competition, discuss current challenges of symbolic regression algorithms and highlight possible improvements for future competitions.
Local Search is a Remarkably Strong Baseline for Neural Architecture Search
Apr 29, 2020

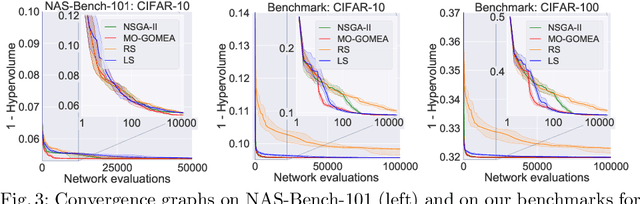
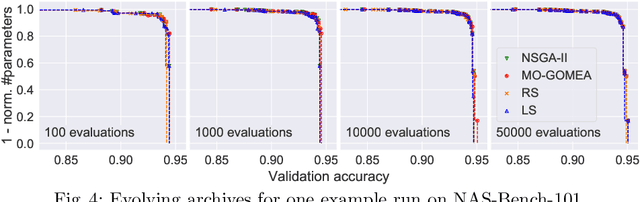
Abstract:Neural Architecture Search (NAS), i.e., the automation of neural network design, has gained much popularity in recent years with increasingly complex search algorithms being proposed. Yet, solid comparisons with simple baselines are often missing. At the same time, recent retrospective studies have found many new algorithms to be no better than random search (RS). In this work we consider, for the first time, a simple Local Search (LS) algorithm for NAS. We particularly consider a multi-objective NAS formulation, with network accuracy and network complexity as two objectives, as understanding the trade-off between these two objectives is arguably the most interesting aspect of NAS. The proposed LS algorithm is compared with RS and two evolutionary algorithms (EAs), as these are often heralded as being ideal for multi-objective optimization. To promote reproducibility, we create and release two benchmark datasets containing 200K saved network evaluations for two established image classification tasks, CIFAR-10 and CIFAR-100. Our benchmarks are designed to be complementary to existing benchmarks, especially in that they are better suited for multi-objective search. We additionally consider a version of the problem with a much larger architecture space. While we find and show that the considered algorithms explore the search space in fundamentally different ways, we also find that LS substantially outperforms RS and even performs nearly as good as state-of-the-art EAs. We believe that this provides strong evidence that LS is truly a competitive baseline for NAS against which new NAS algorithms should be benchmarked.
Surrogate-free machine learning-based organ dose reconstruction for pediatric abdominal radiotherapy
Feb 17, 2020



Abstract:To study radiotherapy-related adverse effects, detailed dose information (3D distribution) is needed for accurate dose-effect modeling. For childhood cancer survivors who underwent radiotherapy in the pre-CT era, only 2D radiographs were acquired, thus 3D dose distributions must be reconstructed. State-of-the-art methods achieve this by using 3D surrogate anatomies. These can however lack personalization and lead to coarse reconstructions. We present and validate a surrogate-free dose reconstruction method based on Machine Learning (ML). Abdominal planning CTs (n=142) of recently-treated childhood cancer patients were gathered, their organs at risk were segmented, and 300 artificial Wilms' tumor plans were sampled automatically. Each artificial plan was automatically emulated on the 142 CTs, resulting in 42,600 3D dose distributions from which dose-volume metrics were derived. Anatomical features were extracted from digitally reconstructed radiographs simulated from the CTs to resemble historical radiographs. Further, patient and radiotherapy plan features typically available from historical treatment records were collected. An evolutionary ML algorithm was then used to link features to dose-volume metrics. Besides 5-fold cross validation, a further evaluation was done on an independent dataset of five CTs each associated with two clinical plans. Cross-validation resulted in mean absolute errors $\leq$0.6 Gy for organs completely inside or outside the field. For organs positioned at the edge of the field, mean absolute errors $\leq$1.7 Gy for $D_{mean}$, $\leq$2.9 Gy for $D_{2cc}$, and $\leq$13% for $V_{5Gy}$ and $V_{10Gy}$, were obtained, without systematic bias. Similar results were found for the independent dataset. To conclude, our novel organ dose reconstruction method is not only accurate, but also efficient, as the setup of a surrogate is no longer needed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge