Ondrej Dyck
Ensemble learning and iterative training (ELIT) machine learning: applications towards uncertainty quantification and automated experiment in atom-resolved microscopy
Jan 22, 2021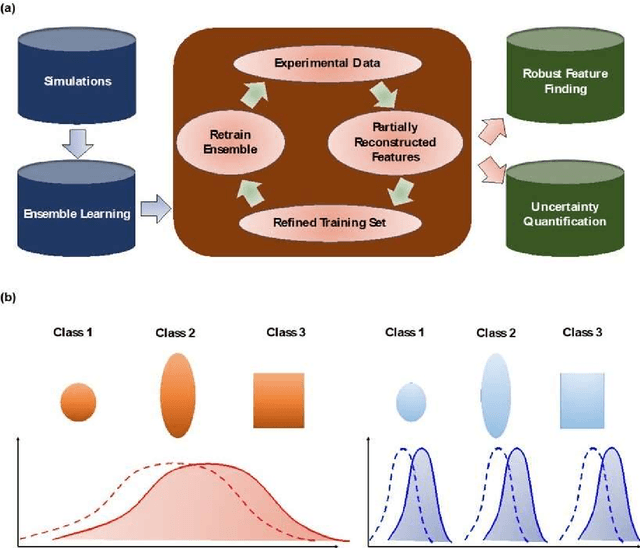
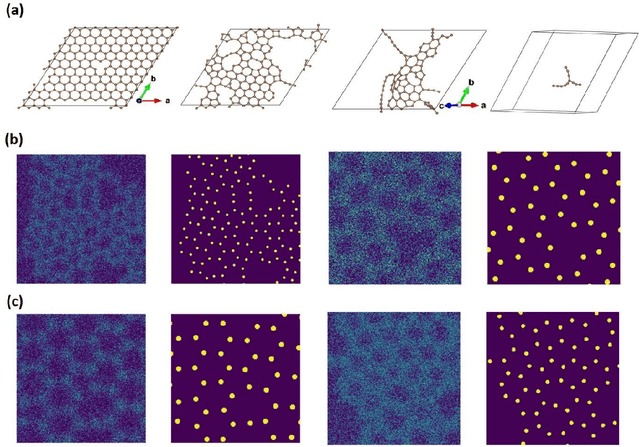
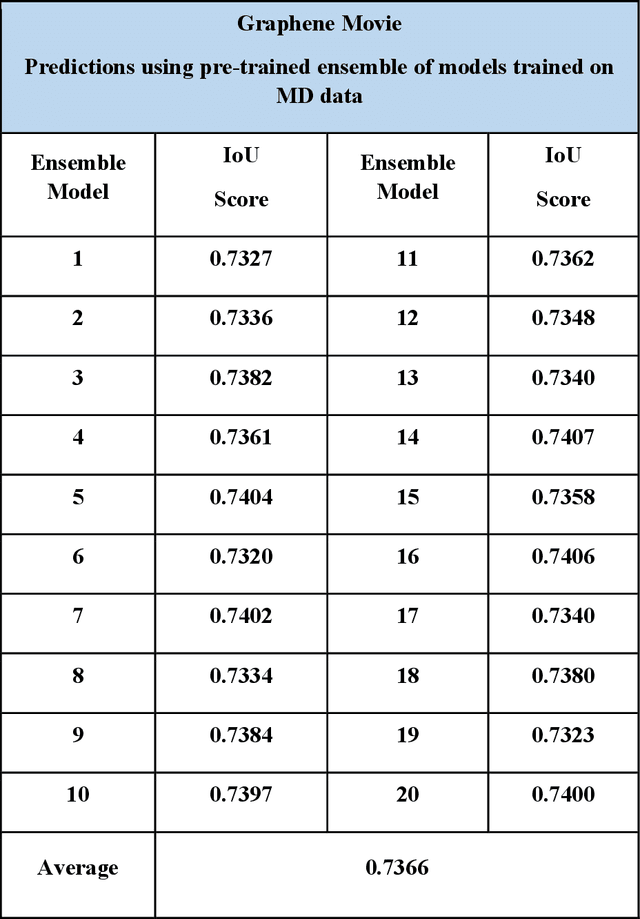
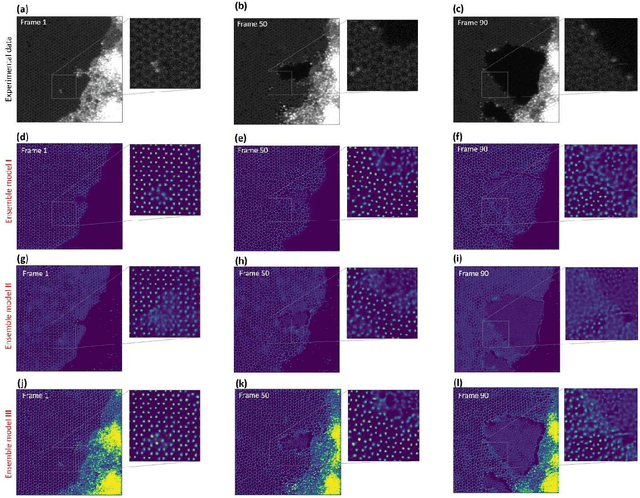
Abstract:Deep learning has emerged as a technique of choice for rapid feature extraction across imaging disciplines, allowing rapid conversion of the data streams to spatial or spatiotemporal arrays of features of interest. However, applications of deep learning in experimental domains are often limited by the out-of-distribution drift between the experiments, where the network trained for one set of imaging conditions becomes sub-optimal for different ones. This limitation is particularly stringent in the quest to have an automated experiment setting, where retraining or transfer learning becomes impractical due to the need for human intervention and associated latencies. Here we explore the reproducibility of deep learning for feature extraction in atom-resolved electron microscopy and introduce workflows based on ensemble learning and iterative training to greatly improve feature detection. This approach both allows incorporating uncertainty quantification into the deep learning analysis and also enables rapid automated experimental workflows where retraining of the network to compensate for out-of-distribution drift due to subtle change in imaging conditions is substituted for a human operator or programmatic selection of networks from the ensemble. This methodology can be further applied to machine learning workflows in other imaging areas including optical and chemical imaging.
Compressed Sensing of Scanning Transmission Electron Microscopy (STEM) on Non-Rectangular Scans
Oct 22, 2018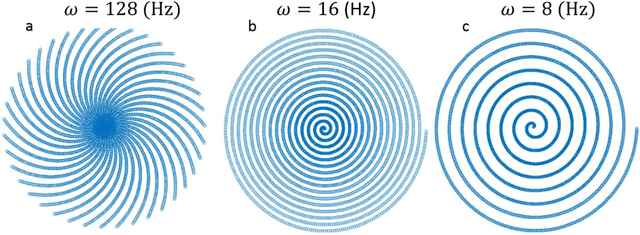
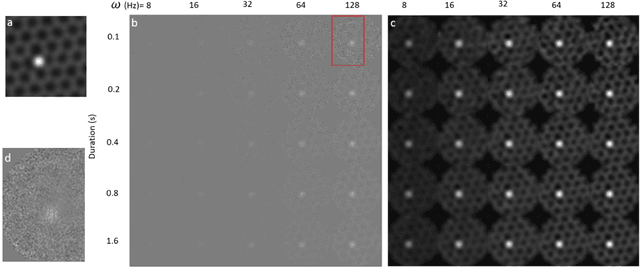
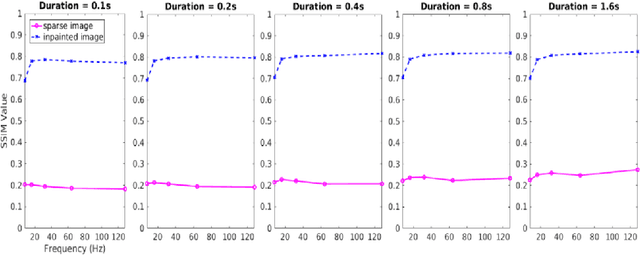
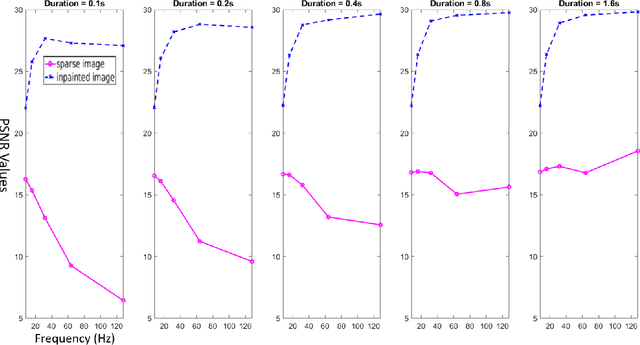
Abstract:Scanning Transmission Electron Microscopy (STEM) has become the main stay for materials characterization on atomic level, with applications ranging from visualization of localized and extended defects to mapping order parameter fields. In the last several years, attention was attracted by potential of STEM to explore beam induced chemical processes and especially manipulating atomic motion, enabling atom-by-atom fabrication. These applications, as well as traditional imaging of beam sensitive materials, necessitate increasing dynamic range of STEM between imaging and manipulation modes, and increasing absolute scanning/imaging speeds, that can be achieved by combining sparse sensing methods with non-rectangular scanning trajectories. Here we developed a general method for real-time reconstruction of sparsely sampled images from high-speed, non-invasive and diverse scanning pathways. This approach is demonstrated on both the synthetic data where ground truth is known and the experimental STEM data. This work lays the foundation for future tasks such as optimal design of dose efficient scanning strategies and real-time adaptive inference and control of e-beam induced atomic fabrication.
Two-Level Structural Sparsity Regularization for Identifying Lattices and Defects in Noisy Images
Sep 01, 2017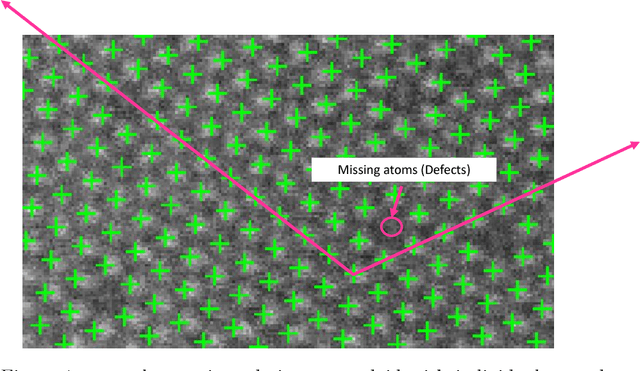
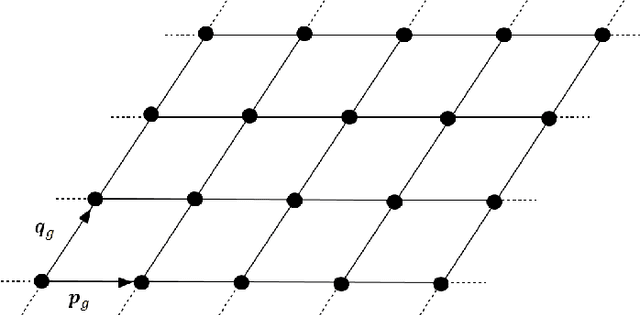
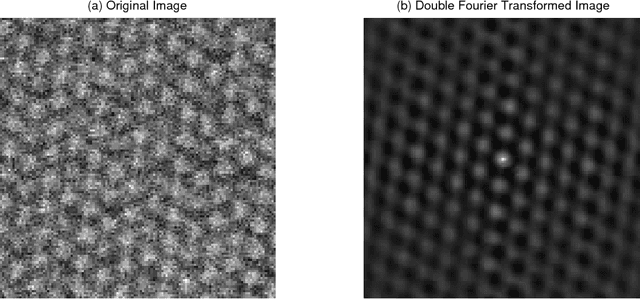
Abstract:This paper presents a regularized regression model with a two-level structural sparsity penalty applied to locate individual atoms in a noisy scanning transmission electron microscopy image (STEM). In crystals, the locations of atoms is symmetric, condensed into a few lattice groups. Therefore, by identifying the underlying lattice in a given image, individual atoms can be accurately located. We propose to formulate the identification of the lattice groups as a sparse group selection problem. Furthermore, real atomic scale images contain defects and vacancies, so atomic identification based solely on a lattice group may result in false positives and false negatives. To minimize error, model includes an individual sparsity regularization in addition to the group sparsity for a within-group selection, which results in a regression model with a two-level sparsity regularization. We propose a modification of the group orthogonal matching pursuit (gOMP) algorithm with a thresholding step to solve the atom finding problem. The convergence and statistical analyses of the proposed algorithm are presented. The proposed algorithm is also evaluated through numerical experiments with simulated images. The applicability of the algorithm on determination of atom structures and identification of imaging distortions and atomic defects was demonstrated using three real STEM images. We believe this is an important step toward automatic phase identification and assignment with the advent of genomic databases for materials.
* Accepted to Annals of Applied Statistics
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge