Nouamane Laanait
Scalable Balanced Training of Conditional Generative Adversarial Neural Networks on Image Data
Feb 21, 2021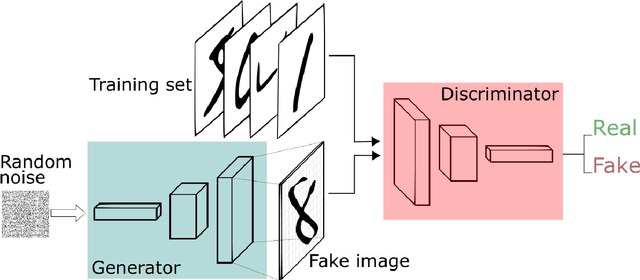
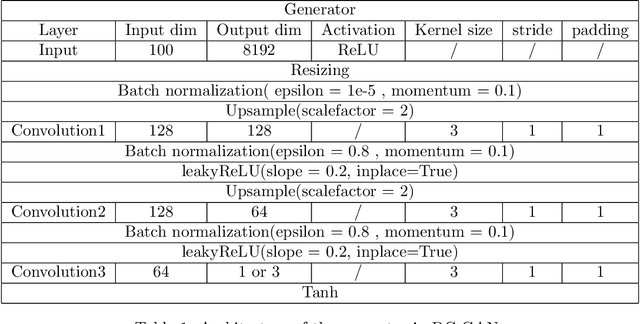
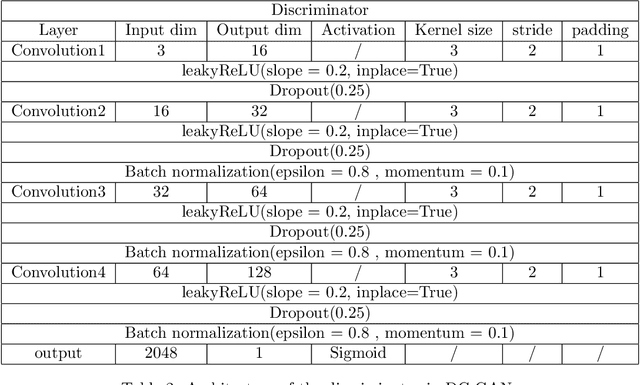
Abstract:We propose a distributed approach to train deep convolutional generative adversarial neural network (DC-CGANs) models. Our method reduces the imbalance between generator and discriminator by partitioning the training data according to data labels, and enhances scalability by performing a parallel training where multiple generators are concurrently trained, each one of them focusing on a single data label. Performance is assessed in terms of inception score and image quality on MNIST, CIFAR10, CIFAR100, and ImageNet1k datasets, showing a significant improvement in comparison to state-of-the-art techniques to training DC-CGANs. Weak scaling is attained on all the four datasets using up to 1,000 processes and 2,000 NVIDIA V100 GPUs on the OLCF supercomputer Summit.
Exascale Deep Learning for Scientific Inverse Problems
Sep 24, 2019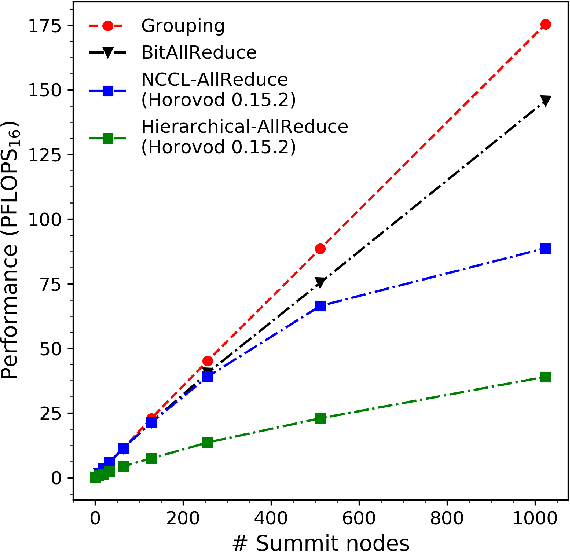
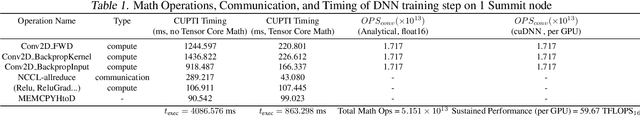
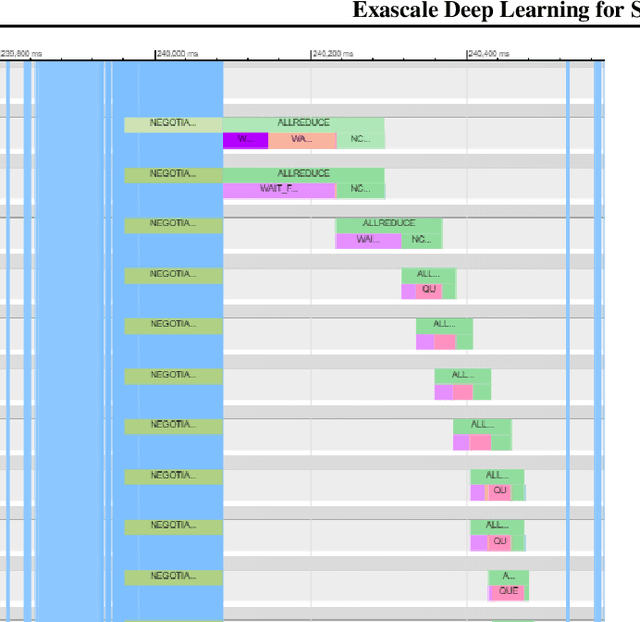

Abstract:We introduce novel communication strategies in synchronous distributed Deep Learning consisting of decentralized gradient reduction orchestration and computational graph-aware grouping of gradient tensors. These new techniques produce an optimal overlap between computation and communication and result in near-linear scaling (0.93) of distributed training up to 27,600 NVIDIA V100 GPUs on the Summit Supercomputer. We demonstrate our gradient reduction techniques in the context of training a Fully Convolutional Neural Network to approximate the solution of a longstanding scientific inverse problem in materials imaging. The efficient distributed training on a dataset size of 0.5 PB, produces a model capable of an atomically-accurate reconstruction of materials, and in the process reaching a peak performance of 2.15(4) EFLOPS$_{16}$.
Reconstruction of 3-D Atomic Distortions from Electron Microscopy with Deep Learning
Feb 19, 2019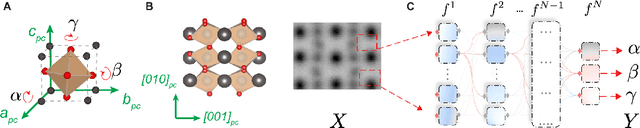
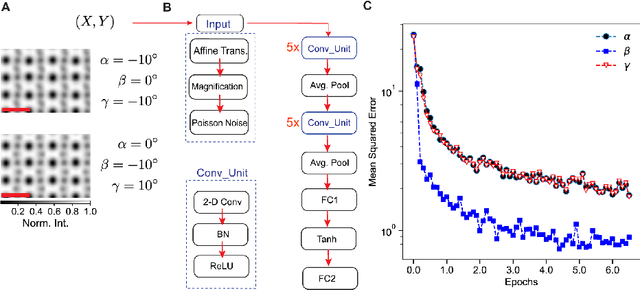
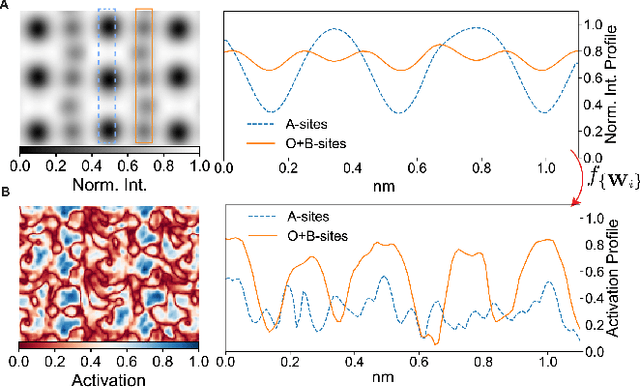
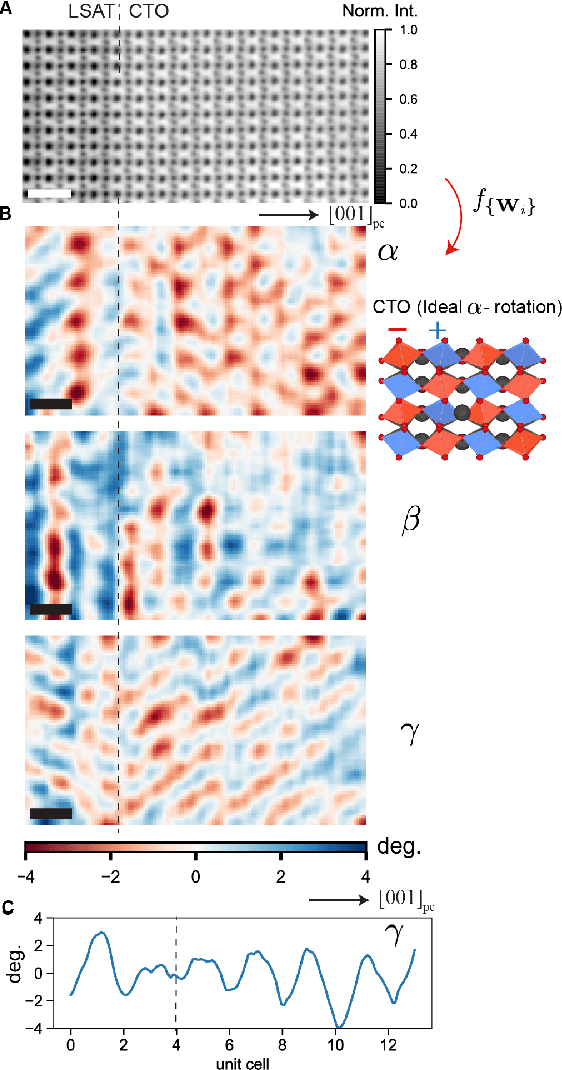
Abstract:Deep learning has demonstrated superb efficacy in processing imaging data, yet its suitability in solving challenging inverse problems in scientific imaging has not been fully explored. Of immense interest is the determination of local material properties from atomically-resolved imaging, such as electron microscopy, where such information is encoded in subtle and complex data signatures, and whose recovery and interpretation necessitate intensive numerical simulations subject to the requirement of near-perfect knowledge of the experimental setup. We demonstrate that an end-to-end deep learning model can successfully recover 3-dimensional atomic distortions of a variety of oxide perovskite materials from a single 2-dimensional experimental scanning transmission electron (STEM) micrograph, in the process resolving a longstanding question in the recovery of 3-D atomic distortions from STEM experiments. Our results indicate that deep learning is a promising approach to efficiently address unsolved inverse problems in scientific imaging and to underpin novel material investigations at atomic resolution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge