Noha A. Yousri
Artificial Intelligence-Based Methods for Precision Medicine: Diabetes Risk Prediction
May 24, 2023Abstract:The rising prevalence of type 2 diabetes mellitus (T2DM) necessitates the development of predictive models for T2DM risk assessment. Artificial intelligence (AI) models are being extensively used for this purpose, but a comprehensive review of their advancements and challenges is lacking. This scoping review analyzes existing literature on AI-based models for T2DM risk prediction. Forty studies were included, mainly published in the past four years. Traditional machine learning models were more prevalent than deep learning models. Electronic health records were the most commonly used data source. Unimodal AI models relying on EHR data were prominent, while only a few utilized multimodal models. Both unimodal and multimodal models showed promising performance, with the latter outperforming the former. Internal validation was common, while external validation was limited. Interpretability methods were reported in half of the studies. Few studies reported novel biomarkers, and open-source code availability was limited. This review provides insights into the current state and limitations of AI-based T2DM risk prediction models and highlights challenges for their development and clinical implementation.
miRNA and Gene Expression based Cancer Classification using Self- Learning and Co-Training Approaches
Jan 18, 2014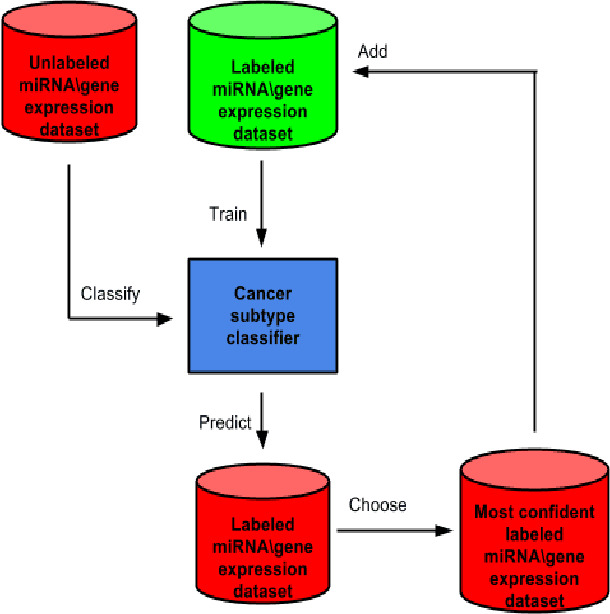
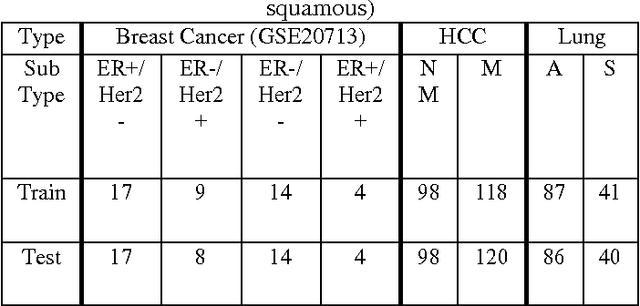

Abstract:miRNA and gene expression profiles have been proved useful for classifying cancer samples. Efficient classifiers have been recently sought and developed. A number of attempts to classify cancer samples using miRNA/gene expression profiles are known in literature. However, the use of semi-supervised learning models have been used recently in bioinformatics, to exploit the huge corpuses of publicly available sets. Using both labeled and unlabeled sets to train sample classifiers, have not been previously considered when gene and miRNA expression sets are used. Moreover, there is a motivation to integrate both miRNA and gene expression for a semi-supervised cancer classification as that provides more information on the characteristics of cancer samples. In this paper, two semi-supervised machine learning approaches, namely self-learning and co-training, are adapted to enhance the quality of cancer sample classification. These approaches exploit the huge public corpuses to enrich the training data. In self-learning, miRNA and gene based classifiers are enhanced independently. While in co-training, both miRNA and gene expression profiles are used simultaneously to provide different views of cancer samples. To our knowledge, it is the first attempt to apply these learning approaches to cancer classification. The approaches were evaluated using breast cancer, hepatocellular carcinoma (HCC) and lung cancer expression sets. Results show up to 20% improvement in F1-measure over Random Forests and SVM classifiers. Co-Training also outperforms Low Density Separation (LDS) approach by around 25% improvement in F1-measure in breast cancer.
Intelligent Hybrid Man-Machine Translation Quality Estimation
Jul 07, 2013



Abstract:Inferring evaluation scores based on human judgments is invaluable compared to using current evaluation metrics which are not suitable for real-time applications e.g. post-editing. However, these judgments are much more expensive to collect especially from expert translators, compared to evaluation based on indicators contrasting source and translation texts. This work introduces a novel approach for quality estimation by combining learnt confidence scores from a probabilistic inference model based on human judgments, with selective linguistic features-based scores, where the proposed inference model infers the credibility of given human ranks to solve the scarcity and inconsistency issues of human judgments. Experimental results, using challenging language-pairs, demonstrate improvement in correlation with human judgments over traditional evaluation metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge