Noah Liniger
Geometric Active Exploration in Markov Decision Processes: the Benefit of Abstraction
Jul 18, 2024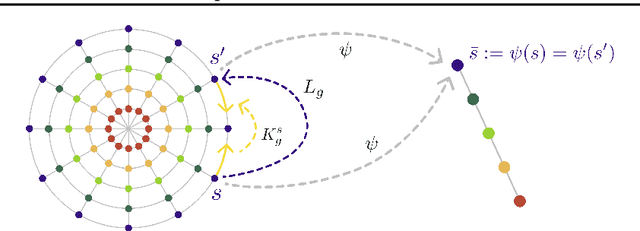
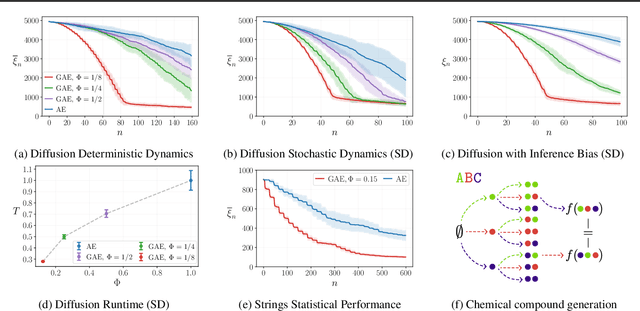
Abstract:How can a scientist use a Reinforcement Learning (RL) algorithm to design experiments over a dynamical system's state space? In the case of finite and Markovian systems, an area called Active Exploration (AE) relaxes the optimization problem of experiments design into Convex RL, a generalization of RL admitting a wider notion of reward. Unfortunately, this framework is currently not scalable and the potential of AE is hindered by the vastness of experiment spaces typical of scientific discovery applications. However, these spaces are often endowed with natural geometries, e.g., permutation invariance in molecular design, that an agent could leverage to improve the statistical and computational efficiency of AE. To achieve this, we bridge AE and MDP homomorphisms, which offer a way to exploit known geometric structures via abstraction. Towards this goal, we make two fundamental contributions: we extend MDP homomorphisms formalism to Convex RL, and we present, to the best of our knowledge, the first analysis that formally captures the benefit of abstraction via homomorphisms on sample efficiency. Ultimately, we propose the Geometric Active Exploration (GAE) algorithm, which we analyse theoretically and experimentally in environments motivated by problems in scientific discovery.
HiPPO-Prophecy: State-Space Models can Provably Learn Dynamical Systems in Context
Jul 12, 2024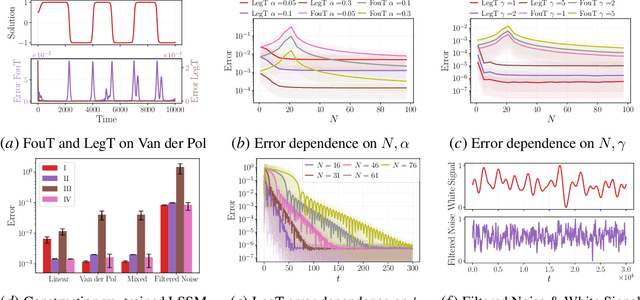
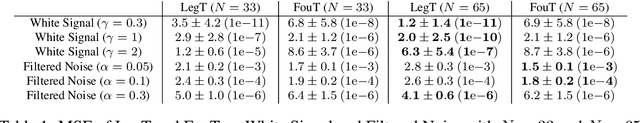
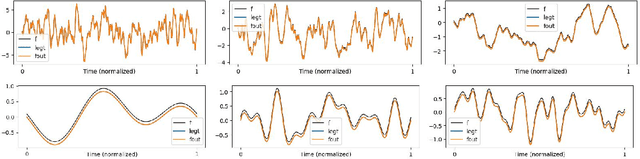
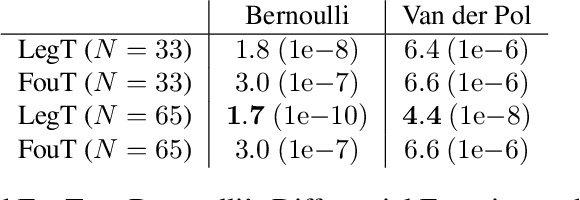
Abstract:This work explores the in-context learning capabilities of State Space Models (SSMs) and presents, to the best of our knowledge, the first theoretical explanation of a possible underlying mechanism. We introduce a novel weight construction for SSMs, enabling them to predict the next state of any dynamical system after observing previous states without parameter fine-tuning. This is accomplished by extending the HiPPO framework to demonstrate that continuous SSMs can approximate the derivative of any input signal. Specifically, we find an explicit weight construction for continuous SSMs and provide an asymptotic error bound on the derivative approximation. The discretization of this continuous SSM subsequently yields a discrete SSM that predicts the next state. Finally, we demonstrate the effectiveness of our parameterization empirically. This work should be an initial step toward understanding how sequence models based on SSMs learn in context.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge