Niloufar Zakariaei
Iterative Flow Matching -- Path Correction and Gradual Refinement for Enhanced Generative Modeling
Feb 26, 2025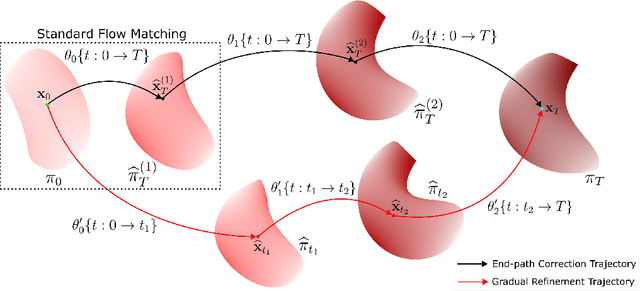
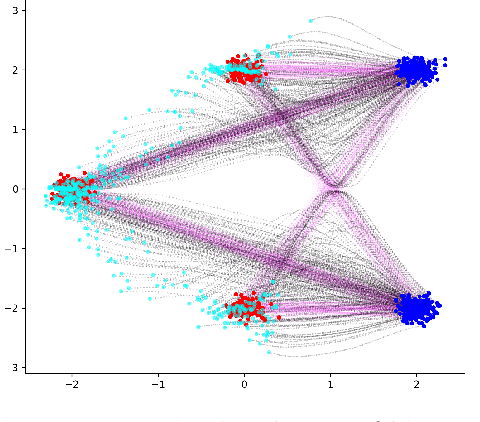
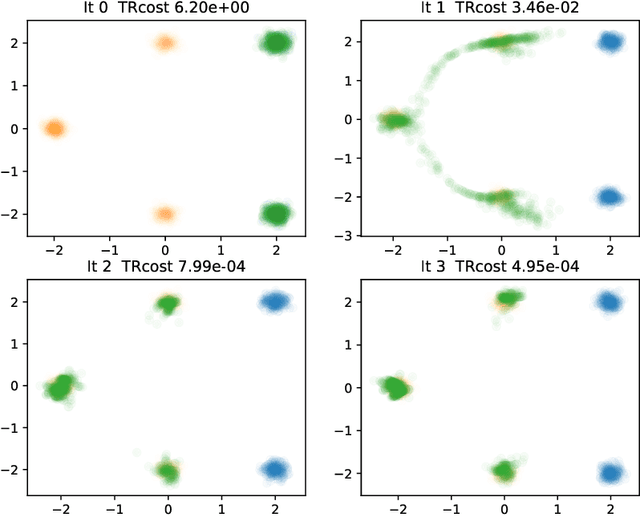
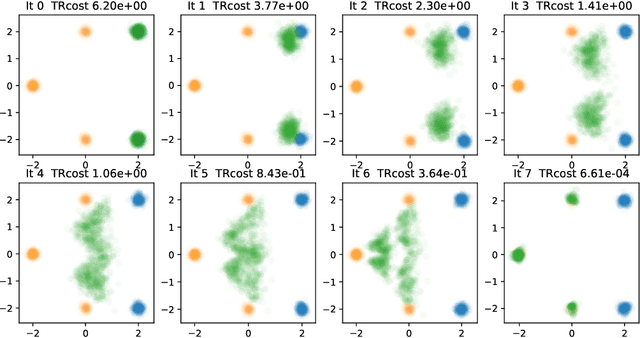
Abstract:Generative models for image generation are now commonly used for a wide variety of applications, ranging from guided image generation for entertainment to solving inverse problems. Nonetheless, training a generator is a non-trivial feat that requires fine-tuning and can lead to so-called hallucinations, that is, the generation of images that are unrealistic. In this work, we explore image generation using flow matching. We explain and demonstrate why flow matching can generate hallucinations, and propose an iterative process to improve the generation process. Our iterative process can be integrated into virtually $\textit{any}$ generative modeling technique, thereby enhancing the performance and robustness of image synthesis systems.
Inversion of Magnetic Data using Learned Dictionaries and Scale Space
Feb 08, 2025Abstract:Magnetic data inversion is an important tool in geophysics, used to infer subsurface magnetic susceptibility distributions from surface magnetic field measurements. This inverse problem is inherently ill-posed, characterized by non-unique solutions, depth ambiguity, and sensitivity to noise. Traditional inversion approaches rely on predefined regularization techniques to stabilize solutions, limiting their adaptability to complex or diverse geological scenarios. In this study, we propose an approach that integrates variable dictionary learning and scale-space methods to address these challenges. Our method employs learned dictionaries, allowing for adaptive representation of complex subsurface features that are difficult to capture with predefined bases. Additionally, we extend classical variational inversion by incorporating multi-scale representations through a scale-space framework, enabling the progressive introduction of structural detail while mitigating overfitting. We implement both fixed and dynamic dictionary learning techniques, with the latter introducing iteration-dependent dictionaries for enhanced flexibility. Using a synthetic dataset to simulate geological scenarios, we demonstrate significant improvements in reconstruction accuracy and robustness compared to conventional variational and dictionary-based methods. Our results highlight the potential of learned dictionaries, especially when coupled with scale-space dynamics, to improve model recovery and noise handling. These findings underscore the promise of our data-driven approach for advance magnetic data inversion and its applications in geophysical exploration, environmental assessment, and mineral prospecting.
Advection Augmented Convolutional Neural Networks
Jun 27, 2024



Abstract:Many problems in physical sciences are characterized by the prediction of space-time sequences. Such problems range from weather prediction to the analysis of disease propagation and video prediction. Modern techniques for the solution of these problems typically combine Convolution Neural Networks (CNN) architecture with a time prediction mechanism. However, oftentimes, such approaches underperform in the long-range propagation of information and lack explainability. In this work, we introduce a physically inspired architecture for the solution of such problems. Namely, we propose to augment CNNs with advection by designing a novel semi-Lagrangian push operator. We show that the proposed operator allows for the non-local transformation of information compared with standard convolutional kernels. We then complement it with Reaction and Diffusion neural components to form a network that mimics the Reaction-Advection-Diffusion equation, in high dimensions. We demonstrate the effectiveness of our network on a number of spatio-temporal datasets that show their merit.
Beyond Conventional Parametric Modeling: Data-Driven Framework for Estimation and Prediction of Time Activity Curves in Dynamic PET Imaging
May 31, 2024Abstract:Dynamic Positron Emission Tomography (dPET) imaging and Time-Activity Curve (TAC) analyses are essential for understanding and quantifying the biodistribution of radiopharmaceuticals over time and space. Traditional compartmental modeling, while foundational, commonly struggles to fully capture the complexities of biological systems, including non-linear dynamics and variability. This study introduces an innovative data-driven neural network-based framework, inspired by Reaction Diffusion systems, designed to address these limitations. Our approach, which adaptively fits TACs from dPET, enables the direct calibration of diffusion coefficients and reaction terms from observed data, offering significant improvements in predictive accuracy and robustness over traditional methods, especially in complex biological scenarios. By more accurately modeling the spatio-temporal dynamics of radiopharmaceuticals, our method advances modeling of pharmacokinetic and pharmacodynamic processes, enabling new possibilities in quantitative nuclear medicine.
MEDDAP: Medical Dataset Enhancement via Diversified Augmentation Pipeline
Mar 26, 2024Abstract:The effectiveness of Deep Neural Networks (DNNs) heavily relies on the abundance and accuracy of available training data. However, collecting and annotating data on a large scale is often both costly and time-intensive, particularly in medical cases where practitioners are already occupied with their duties. Moreover, ensuring that the model remains robust across various scenarios of image capture is crucial in medical domains, especially when dealing with ultrasound images that vary based on the settings of different devices and the manual operation of the transducer. To address this challenge, we introduce a novel pipeline called MEDDAP, which leverages Stable Diffusion (SD) models to augment existing small datasets by automatically generating new informative labeled samples. Pretrained checkpoints for SD are typically based on natural images, and training them for medical images requires significant GPU resources due to their heavy parameters. To overcome this challenge, we introduce USLoRA (Ultrasound Low-Rank Adaptation), a novel fine-tuning method tailored specifically for ultrasound applications. USLoRA allows for selective fine-tuning of weights within SD, requiring fewer than 0.1\% of parameters compared to fully fine-tuning only the UNet portion of SD. To enhance dataset diversity, we incorporate different adjectives into the generation process prompts, thereby desensitizing the classifiers to intensity changes across different images. This approach is inspired by clinicians' decision-making processes regarding breast tumors, where tumor shape often plays a more crucial role than intensity. In conclusion, our pipeline not only outperforms classifiers trained on the original dataset but also demonstrates superior performance when encountering unseen datasets. The source code is available at https://github.com/yasamin-med/MEDDAP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge