Nicki Skafte Detlefsen
Is an encoder within reach?
Jun 03, 2022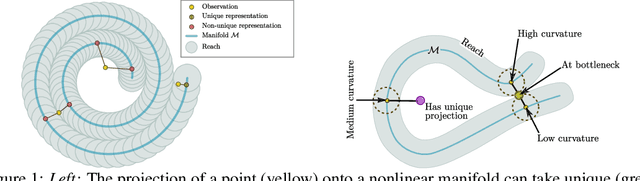

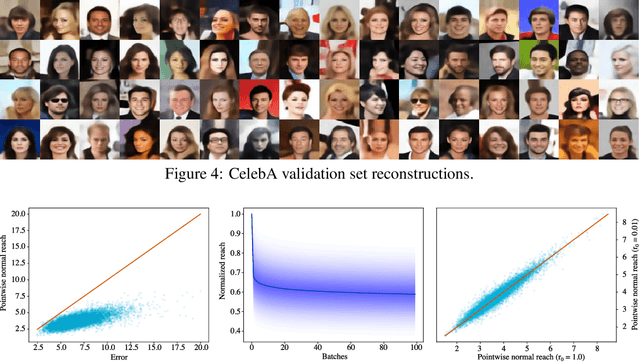

Abstract:The encoder network of an autoencoder is an approximation of the nearest point projection onto the manifold spanned by the decoder. A concern with this approximation is that, while the output of the encoder is always unique, the projection can possibly have infinitely many values. This implies that the latent representations learned by the autoencoder can be misleading. Borrowing from geometric measure theory, we introduce the idea of using the reach of the manifold spanned by the decoder to determine if an optimal encoder exists for a given dataset and decoder. We develop a local generalization of this reach and propose a numerical estimator thereof. We demonstrate that this allows us to determine which observations can be expected to have a unique, and thereby trustworthy, latent representation. As our local reach estimator is differentiable, we investigate its usage as a regularizer and show that this leads to learned manifolds for which projections are more often unique than without regularization.
What is a meaningful representation of protein sequences?
Nov 28, 2020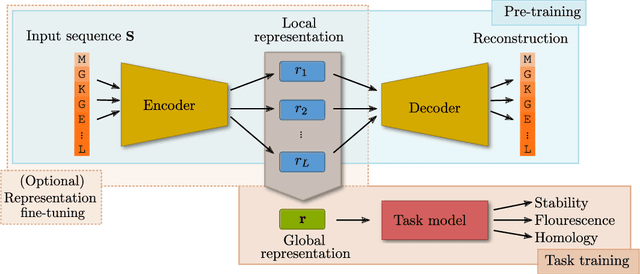
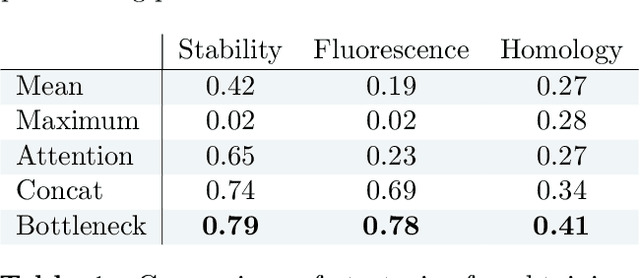
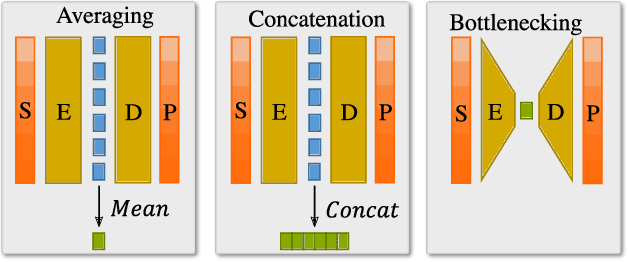
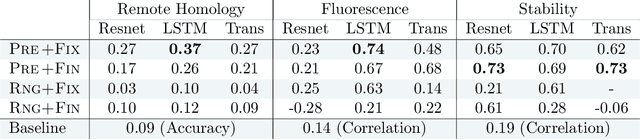
Abstract:How we choose to represent our data has a fundamental impact on our ability to subsequently extract information from them. Machine learning promises to automatically determine efficient representations from large unstructured datasets, such as those arising in biology. However, empirical evidence suggests that seemingly minor changes to these machine learning models yield drastically different data representations that result in different biological interpretations of data. This begs the question of what even constitutes the most meaningful representation. Here, we approach this question for representations of protein sequences, which have received considerable attention in the recent literature. We explore two key contexts in which representations naturally arise: transfer learning and interpretable learning. In the first context, we demonstrate that several contemporary practices yield suboptimal performance, and in the latter we demonstrate that taking representation geometry into account significantly improves interpretability and lets the models reveal biological information that is otherwise obscured.
Explicit Disentanglement of Appearance and Perspective in Generative Models
Jun 11, 2019



Abstract:Disentangled representation learning finds compact, independent and easy-to-interpret factors of the data. Learning such has been shown to require an inductive bias, which we explicitly encode in a generative model of images. Specifically, we propose a model with two latent spaces: one that represents spatial transformations of the input data, and another that represents the transformed data. We find that the latter naturally captures the intrinsic appearance of the data. To realize the generative model, we propose a Variationally Inferred Transformational Autoencoder (VITAE) that incorporates a spatial transformer into a variational autoencoder. We show how to perform inference in the model efficiently by carefully designing the encoders and restricting the transformation class to be diffeomorphic. Empirically, our model separates the visual style from digit type on MNIST, and separates shape and pose in images of the human body.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge