Navid Ziaei
A Combined Channel Approach for Decoding Intracranial EEG Signals: Enhancing Accuracy through Spatial Information Integration
Dec 09, 2024



Abstract:Intracranial EEG (iEEG) recording, characterized by high spatial and temporal resolution and superior signal-to-noise ratio (SNR), enables the development of precise brain-computer interface (BCI) systems for neural decoding. However, the invasive nature of the procedure significantly limits the availability of iEEG datasets in terms of both the number of participants and the duration of recorded sessions. To address this limitation, we propose a single-participant machine learning model optimized for decoding iEEG signals. The model employs 18 key features and operates in two modes: best channel and combined channel. The combined channel mode integrates spatial information from multiple brain regions, leading to superior classification performance. Evaluations across three datasets -- Music Reconstruction, Audio Visual, and AJILE12 -- demonstrate that the combined channel mode consistently outperforms the best channel mode across all classifiers. In the best-performing cases, Random Forest achieved an F1 score of 0.81 +/- 0.05 in the Music Reconstruction dataset and 0.82 +/- 0.10 in the Audio Visual dataset, while XGBoost achieved an F1 score of 0.84 +/- 0.08 in the AJILE12 dataset. Furthermore, the analysis of brain region contributions in the combined channel mode revealed that the model identifies relevant brain regions aligned with physiological expectations for each task and effectively combines data from electrodes in these regions to achieve high performance. These findings highlight the potential of integrating spatial information across brain regions to improve task decoding, offering new avenues for advancing BCI systems and neurotechnological applications.
Latent Variable Double Gaussian Process Model for Decoding Complex Neural Data
May 08, 2024Abstract:Non-parametric models, such as Gaussian Processes (GP), show promising results in the analysis of complex data. Their applications in neuroscience data have recently gained traction. In this research, we introduce a novel neural decoder model built upon GP models. The core idea is that two GPs generate neural data and their associated labels using a set of low-dimensional latent variables. Under this modeling assumption, the latent variables represent the underlying manifold or essential features present in the neural data. When GPs are trained, the latent variable can be inferred from neural data to decode the labels with a high accuracy. We demonstrate an application of this decoder model in a verbal memory experiment dataset and show that the decoder accuracy in predicting stimulus significantly surpasses the state-of-the-art decoder models. The preceding performance of this model highlights the importance of utilizing non-parametric models in the analysis of neuroscience data.
A Discriminative Bayesian Gaussian Process Latent Variable Model for High-Dimensional Data
Jan 29, 2024



Abstract:Extracting meaningful information from high-dimensional data poses a formidable modeling challenge, particularly when the data is obscured by noise or represented through different modalities. In this research, we propose a novel non-parametric modeling approach, leveraging the Gaussian Process (GP), to characterize high-dimensional data by mapping it to a latent low-dimensional manifold. This model, named the Latent Discriminative Generative Decoder (LDGD), utilizes both the data (or its features) and associated labels (such as category or stimulus) in the manifold discovery process. To infer the latent variables, we derive a Bayesian solution, allowing LDGD to effectively capture inherent uncertainties in the data while enhancing the model's predictive accuracy and robustness. We demonstrate the application of LDGD on both synthetic and benchmark datasets. Not only does LDGD infer the manifold accurately, but its prediction accuracy in anticipating labels surpasses state-of-the-art approaches. We have introduced inducing points to reduce the computational complexity of Gaussian Processes (GPs) for large datasets. This enhancement facilitates batch training, allowing for more efficient processing and scalability in handling extensive data collections. Additionally, we illustrate that LDGD achieves higher accuracy in predicting labels and operates effectively with a limited training dataset, underscoring its efficiency and effectiveness in scenarios where data availability is constrained. These attributes set the stage for the development of non-parametric modeling approaches in the analysis of high-dimensional data; especially in fields where data are both high-dimensional and complex.
Bayesian Time-Series Classifier for Decoding Simple Visual Stimuli from Intracranial Neural Activity
Jul 28, 2023Abstract:Understanding how external stimuli are encoded in distributed neural activity is of significant interest in clinical and basic neuroscience. To address this need, it is essential to develop analytical tools capable of handling limited data and the intrinsic stochasticity present in neural data. In this study, we propose a straightforward Bayesian time series classifier (BTsC) model that tackles these challenges whilst maintaining a high level of interpretability. We demonstrate the classification capabilities of this approach by utilizing neural data to decode colors in a visual task. The model exhibits consistent and reliable average performance of 75.55% on 4 patients' dataset, improving upon state-of-the-art machine learning techniques by about 3.0 percent. In addition to its high classification accuracy, the proposed BTsC model provides interpretable results, making the technique a valuable tool to study neural activity in various tasks and categories. The proposed solution can be applied to neural data recorded in various tasks, where there is a need for interpretable results and accurate classification accuracy.
Reverse Survival Model (RSM): A Pipeline for Explaining Predictions of Deep Survival Models
Oct 27, 2022Abstract:The aim of survival analysis in healthcare is to estimate the probability of occurrence of an event, such as a patient's death in an intensive care unit (ICU). Recent developments in deep neural networks (DNNs) for survival analysis show the superiority of these models in comparison with other well-known models in survival analysis applications. Ensuring the reliability and explainability of deep survival models deployed in healthcare is a necessity. Since DNN models often behave like a black box, their predictions might not be easily trusted by clinicians, especially when predictions are contrary to a physician's opinion. A deep survival model that explains and justifies its decision-making process could potentially gain the trust of clinicians. In this research, we propose the reverse survival model (RSM) framework that provides detailed insights into the decision-making process of survival models. For each patient of interest, RSM can extract similar patients from a dataset and rank them based on the most relevant features that deep survival models rely on for their predictions.
Survival Seq2Seq: A Survival Model based on Sequence to Sequence Architecture
Apr 09, 2022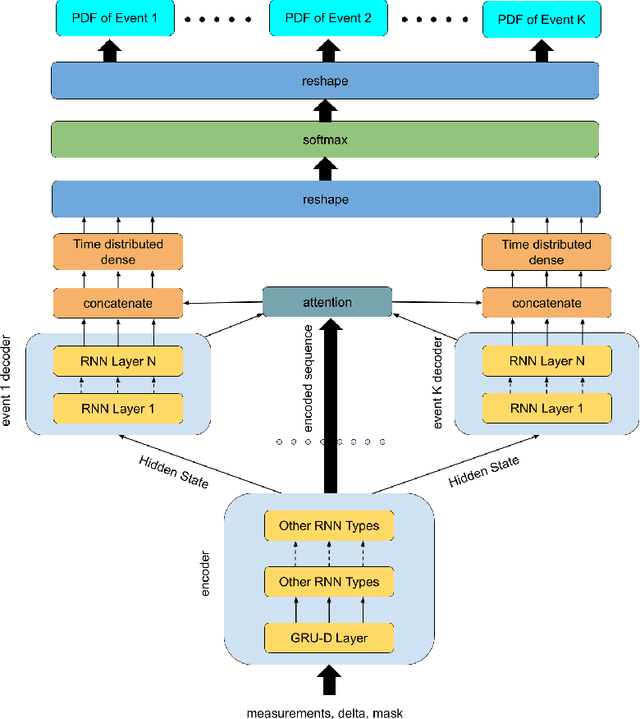
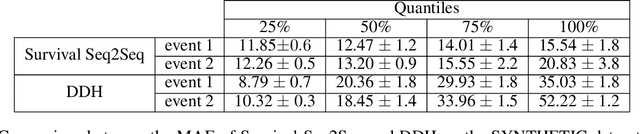
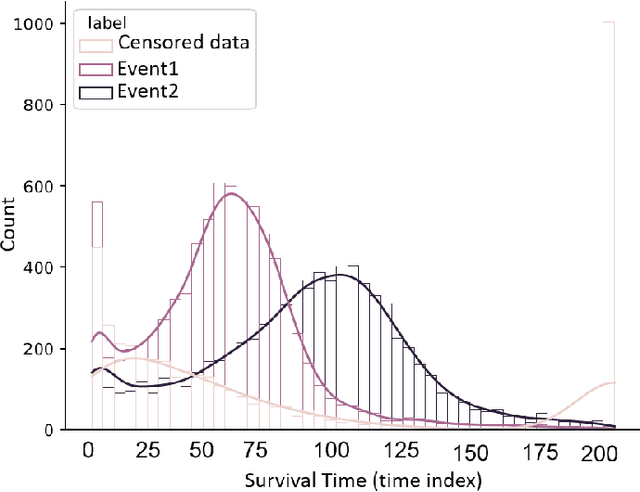
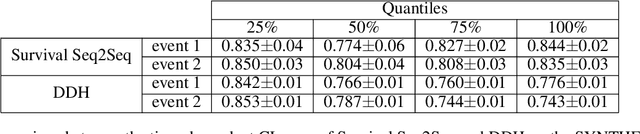
Abstract:This paper introduces a novel non-parametric deep model for estimating time-to-event (survival analysis) in presence of censored data and competing risks. The model is designed based on the sequence-to-sequence (Seq2Seq) architecture, therefore we name it Survival Seq2Seq. The first recurrent neural network (RNN) layer of the encoder of our model is made up of Gated Recurrent Unit with Decay (GRU-D) cells. These cells have the ability to effectively impute not-missing-at-random values of longitudinal datasets with very high missing rates, such as electronic health records (EHRs). The decoder of Survival Seq2Seq generates a probability distribution function (PDF) for each competing risk without assuming any prior distribution for the risks. Taking advantage of RNN cells, the decoder is able to generate smooth and virtually spike-free PDFs. This is beyond the capability of existing non-parametric deep models for survival analysis. Training results on synthetic and medical datasets prove that Survival Seq2Seq surpasses other existing deep survival models in terms of the accuracy of predictions and the quality of generated PDFs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge