Ali Yousefi
Latent Variable Double Gaussian Process Model for Decoding Complex Neural Data
May 08, 2024Abstract:Non-parametric models, such as Gaussian Processes (GP), show promising results in the analysis of complex data. Their applications in neuroscience data have recently gained traction. In this research, we introduce a novel neural decoder model built upon GP models. The core idea is that two GPs generate neural data and their associated labels using a set of low-dimensional latent variables. Under this modeling assumption, the latent variables represent the underlying manifold or essential features present in the neural data. When GPs are trained, the latent variable can be inferred from neural data to decode the labels with a high accuracy. We demonstrate an application of this decoder model in a verbal memory experiment dataset and show that the decoder accuracy in predicting stimulus significantly surpasses the state-of-the-art decoder models. The preceding performance of this model highlights the importance of utilizing non-parametric models in the analysis of neuroscience data.
A Discriminative Bayesian Gaussian Process Latent Variable Model for High-Dimensional Data
Jan 29, 2024



Abstract:Extracting meaningful information from high-dimensional data poses a formidable modeling challenge, particularly when the data is obscured by noise or represented through different modalities. In this research, we propose a novel non-parametric modeling approach, leveraging the Gaussian Process (GP), to characterize high-dimensional data by mapping it to a latent low-dimensional manifold. This model, named the Latent Discriminative Generative Decoder (LDGD), utilizes both the data (or its features) and associated labels (such as category or stimulus) in the manifold discovery process. To infer the latent variables, we derive a Bayesian solution, allowing LDGD to effectively capture inherent uncertainties in the data while enhancing the model's predictive accuracy and robustness. We demonstrate the application of LDGD on both synthetic and benchmark datasets. Not only does LDGD infer the manifold accurately, but its prediction accuracy in anticipating labels surpasses state-of-the-art approaches. We have introduced inducing points to reduce the computational complexity of Gaussian Processes (GPs) for large datasets. This enhancement facilitates batch training, allowing for more efficient processing and scalability in handling extensive data collections. Additionally, we illustrate that LDGD achieves higher accuracy in predicting labels and operates effectively with a limited training dataset, underscoring its efficiency and effectiveness in scenarios where data availability is constrained. These attributes set the stage for the development of non-parametric modeling approaches in the analysis of high-dimensional data; especially in fields where data are both high-dimensional and complex.
Bayesian Time-Series Classifier for Decoding Simple Visual Stimuli from Intracranial Neural Activity
Jul 28, 2023Abstract:Understanding how external stimuli are encoded in distributed neural activity is of significant interest in clinical and basic neuroscience. To address this need, it is essential to develop analytical tools capable of handling limited data and the intrinsic stochasticity present in neural data. In this study, we propose a straightforward Bayesian time series classifier (BTsC) model that tackles these challenges whilst maintaining a high level of interpretability. We demonstrate the classification capabilities of this approach by utilizing neural data to decode colors in a visual task. The model exhibits consistent and reliable average performance of 75.55% on 4 patients' dataset, improving upon state-of-the-art machine learning techniques by about 3.0 percent. In addition to its high classification accuracy, the proposed BTsC model provides interpretable results, making the technique a valuable tool to study neural activity in various tasks and categories. The proposed solution can be applied to neural data recorded in various tasks, where there is a need for interpretable results and accurate classification accuracy.
Deep Discriminative Direct Decoders for High-dimensional Time-series Analysis
May 22, 2022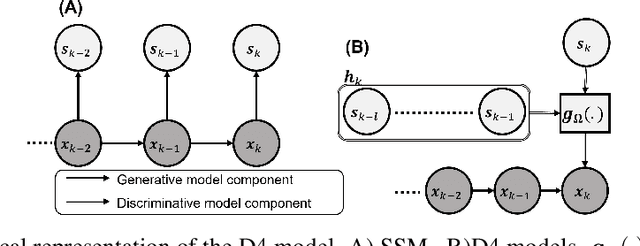
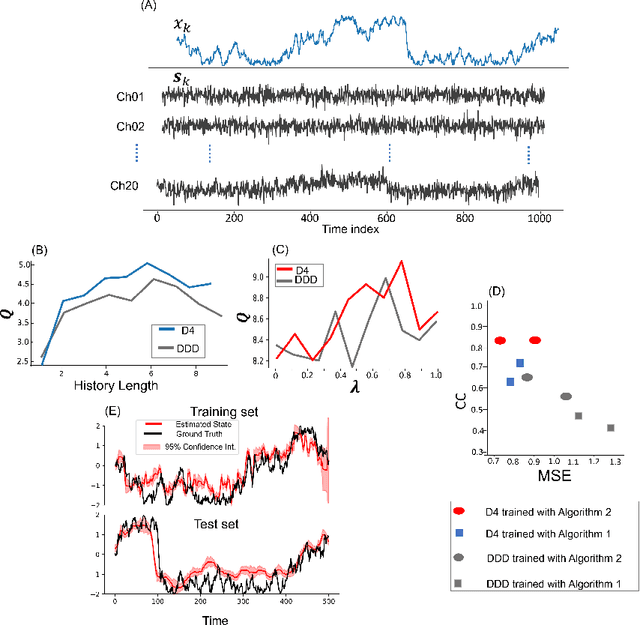
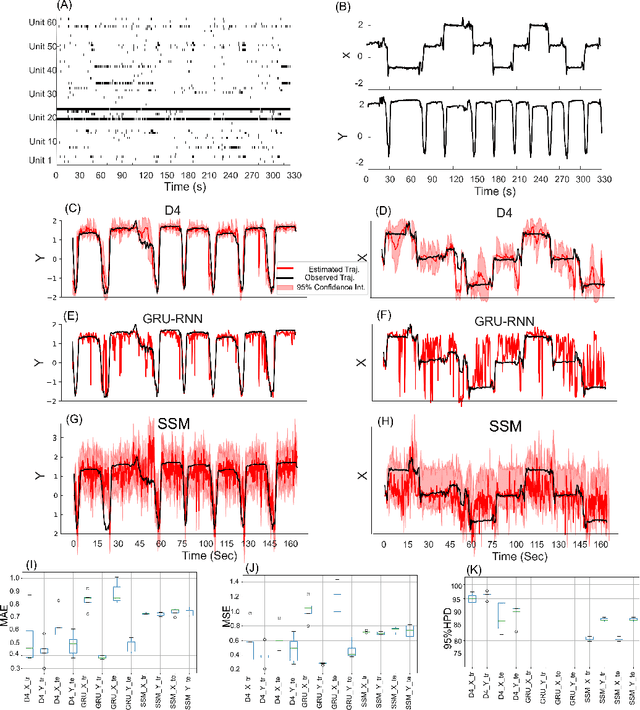
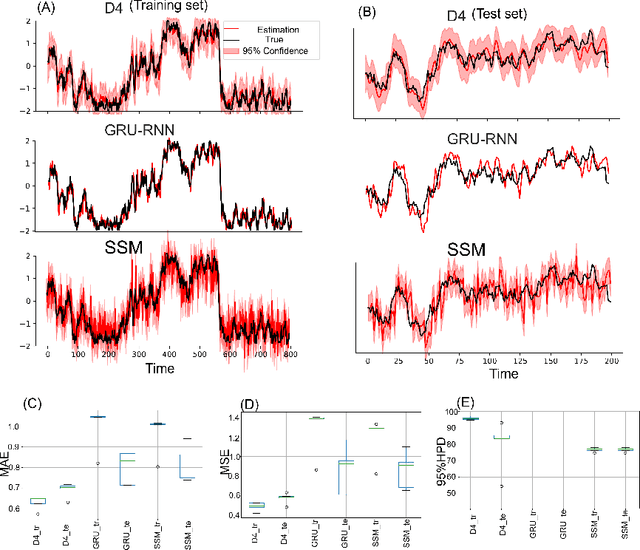
Abstract:Dynamical latent variable modeling has been significantly invested over the last couple of decades with established solutions encompassing generative processes like the state-space model (SSM) and discriminative processes like a recurrent or a deep neural network (DNN). These solutions are powerful tools with promising results; however, surprisingly they were never put together in a unified model to analyze complex multivariate time-series data. A very recent modeling approach, called the direct discriminative decoder (DDD) model, proposes a principal solution to combine SMM and DNN models, with promising results in decoding underlying latent processes, e.g. rat movement trajectory, through high-dimensional neural recordings. The DDD consists of a) a state transition process, as per the classical dynamical models, and b) a discriminative process, like DNN, in which the conditional distribution of states is defined as a function of the current observations and their recent history. Despite promising results of the DDD model, no training solutions, in the context of DNN, have been utilized for this model. Here, we propose how DNN parameters along with an optimal history term can be simultaneously estimated as a part of the DDD model. We use the D4 abbreviation for a DDD with a DNN as its discriminative process. We showed the D4 decoding performance in both simulation and (relatively) high-dimensional neural data. In both datasets, D4 performance surpasses the state-of-art decoding solutions, including those of SSM and DNNs. The key success of DDD and potentially D4 is efficient utilization of the recent history of observation along with the state-process that carries long-term information, which is not addressed in either SSM or DNN solutions. We argue that D4 can be a powerful tool for the analysis of high-dimensional time-series data.
Latent Factor Decomposition Model: Applications for Questionnaire Data
Apr 30, 2021



Abstract:The analysis of clinical questionnaire data comes with many inherent challenges. These challenges include the handling of data with missing fields, as well as the overall interpretation of a dataset with many fields of different scales and forms. While numerous methods have been developed to address these challenges, they are often not robust, statistically sound, or easily interpretable. Here, we propose a latent factor modeling framework that extends the principal component analysis for both categorical and quantitative data with missing elements. The model simultaneously provides the principal components (basis) and each patients' projections on these bases in a latent space. We show an application of our modeling framework through Irritable Bowel Syndrome (IBS) symptoms, where we find correlations between these projections and other standardized patient symptom scales. This latent factor model can be easily applied to different clinical questionnaire datasets for clustering analysis and interpretable inference.
Foot anthropometry device and single object image thresholding
Jul 10, 2017



Abstract:This paper introduces a device, algorithm and graphical user interface to obtain anthropometric measurements of foot. Presented device facilitates obtaining scale of image and image processing by taking one image from side foot and underfoot simultaneously. Introduced image processing algorithm minimizes a noise criterion, which is suitable for object detection in single object images and outperforms famous image thresholding methods when lighting condition is poor. Performance of image-based method is compared to manual method. Image-based measurements of underfoot in average was 4mm less than actual measures. Mean absolute error of underfoot length was 1.6mm, however length obtained from side foot had 4.4mm mean absolute error. Furthermore, based on t-test and f-test results, no significant difference between manual and image-based anthropometry observed. In order to maintain anthropometry process performance in different situations user interface designed for handling changes in light conditions and altering speed of the algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge