Nathan Baker
How Much Chemistry Does a Deep Neural Network Need to Know to Make Accurate Predictions?
Mar 18, 2018
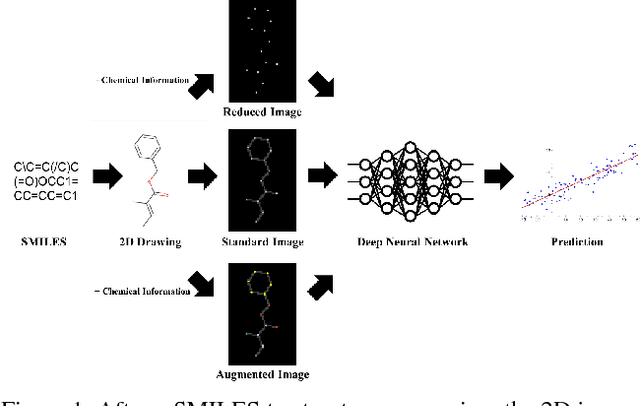
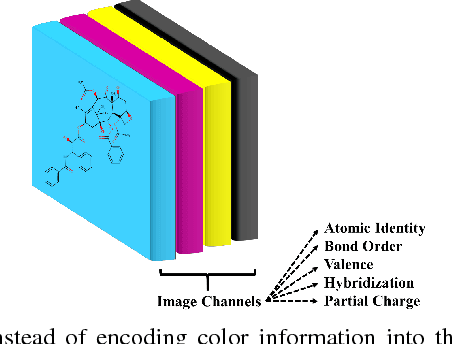
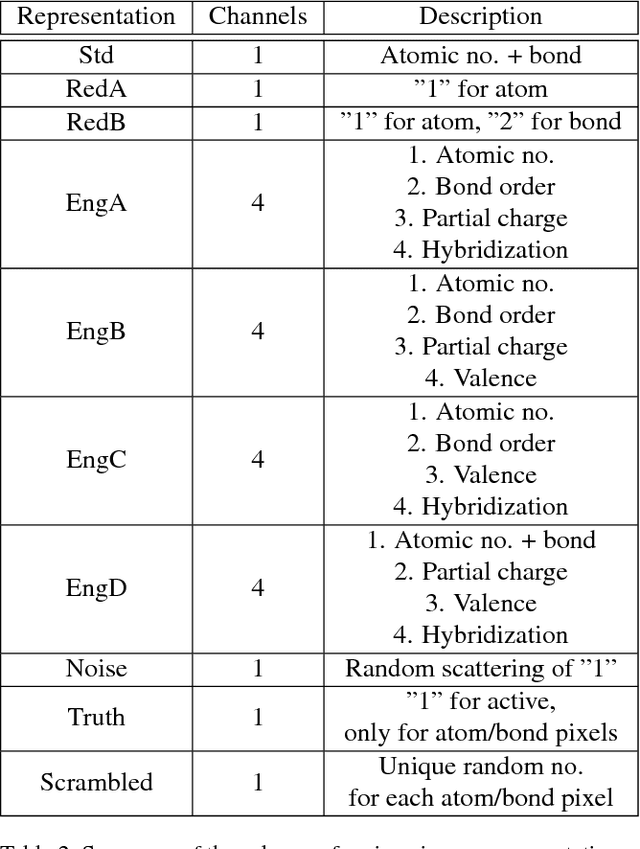
Abstract:The meteoric rise of deep learning models in computer vision research, having achieved human-level accuracy in image recognition tasks is firm evidence of the impact of representation learning of deep neural networks. In the chemistry domain, recent advances have also led to the development of similar CNN models, such as Chemception, that is trained to predict chemical properties using images of molecular drawings. In this work, we investigate the effects of systematically removing and adding localized domain-specific information to the image channels of the training data. By augmenting images with only 3 additional basic information, and without introducing any architectural changes, we demonstrate that an augmented Chemception (AugChemception) outperforms the original model in the prediction of toxicity, activity, and solvation free energy. Then, by altering the information content in the images, and examining the resulting model's performance, we also identify two distinct learning patterns in predicting toxicity/activity as compared to solvation free energy. These patterns suggest that Chemception is learning about its tasks in the manner that is consistent with established knowledge. Thus, our work demonstrates that advanced chemical knowledge is not a pre-requisite for deep learning models to accurately predict complex chemical properties.
Chemception: A Deep Neural Network with Minimal Chemistry Knowledge Matches the Performance of Expert-developed QSAR/QSPR Models
Jun 20, 2017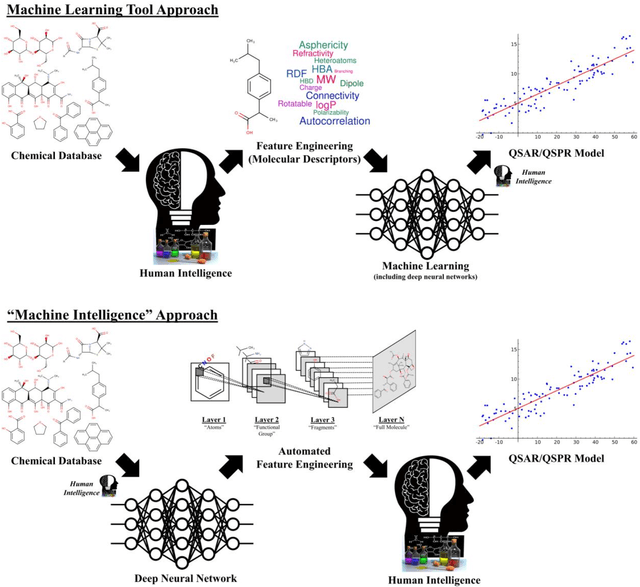
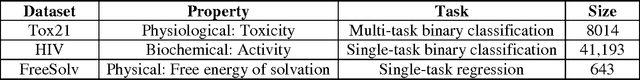
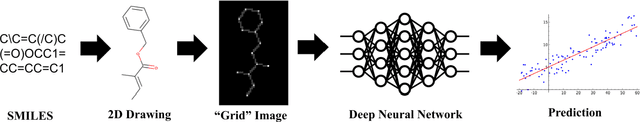
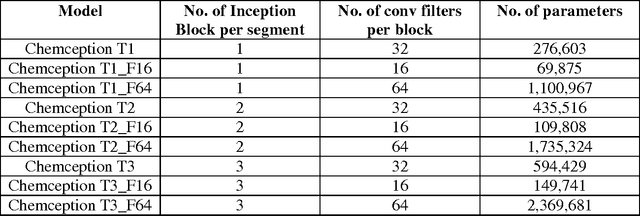
Abstract:In the last few years, we have seen the transformative impact of deep learning in many applications, particularly in speech recognition and computer vision. Inspired by Google's Inception-ResNet deep convolutional neural network (CNN) for image classification, we have developed "Chemception", a deep CNN for the prediction of chemical properties, using just the images of 2D drawings of molecules. We develop Chemception without providing any additional explicit chemistry knowledge, such as basic concepts like periodicity, or advanced features like molecular descriptors and fingerprints. We then show how Chemception can serve as a general-purpose neural network architecture for predicting toxicity, activity, and solvation properties when trained on a modest database of 600 to 40,000 compounds. When compared to multi-layer perceptron (MLP) deep neural networks trained with ECFP fingerprints, Chemception slightly outperforms in activity and solvation prediction and slightly underperforms in toxicity prediction. Having matched the performance of expert-developed QSAR/QSPR deep learning models, our work demonstrates the plausibility of using deep neural networks to assist in computational chemistry research, where the feature engineering process is performed primarily by a deep learning algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge