Nabeel Abdur Rehman
Population-aware Hierarchical Bayesian Domain Adaptation via Multiple-component Invariant Learning
Sep 13, 2019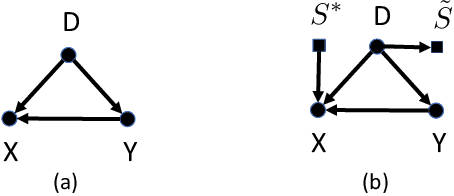
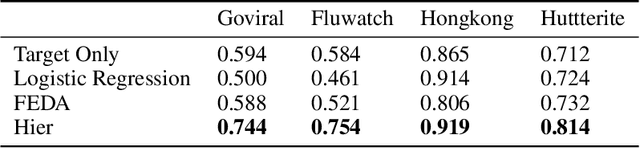
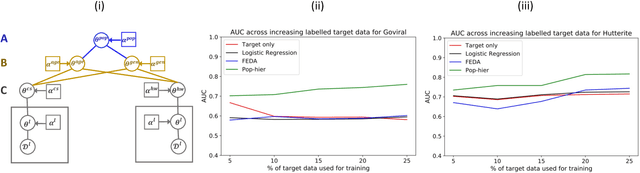
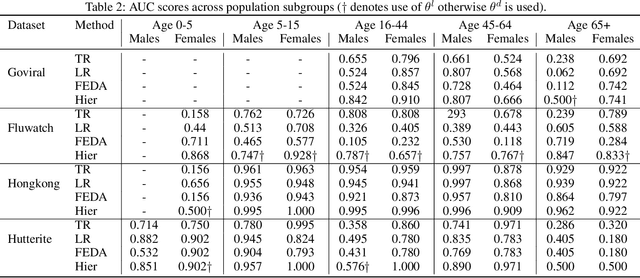
Abstract:While machine learning is rapidly being developed and deployed in health settings such as influenza prediction, there are critical challenges in using data from one environment in another due to variability in features; even within disease labels there can be differences (e.g. "fever" may mean something different reported in a doctor's office versus in an online app). Moreover, models are often built on passive, observational data which contain different distributions of population subgroups (e.g. men or women). Thus, there are two forms of instability between environments in this observational transport problem. We first harness knowledge from health to conceptualize the underlying causal structure of this problem in a health outcome prediction task. Based on sources of stability in the model, we posit that for human-sourced data and health prediction tasks we can combine environment and population information in a novel population-aware hierarchical Bayesian domain adaptation framework that harnesses multiple invariant components through population attributes when needed. We study the conditions under which invariant learning fails, leading to reliance on the environment-specific attributes. Experimental results for an influenza prediction task on four datasets gathered from different contexts show the model can improve prediction in the case of largely unlabelled target data from a new environment and different constituent population, by harnessing both environment and population invariant information. This work represents a novel, principled way to address a critical challenge by blending domain (health) knowledge and algorithmic innovation. The proposed approach will have a significant impact in many social settings wherein who and where the data comes from matters.
Deep Landscape Features for Improving Vector-borne Disease Prediction
Apr 03, 2019



Abstract:The global population at risk of mosquito-borne diseases such as dengue, yellow fever, chikungunya and Zika is expanding. Infectious disease models commonly incorporate environmental measures like temperature and precipitation. Given increasing availability of high-resolution satellite imagery, here we consider including landscape features from satellite imagery into infectious disease prediction models. To do so, we implement a Convolutional Neural Network (CNN) model trained on Imagenet data and labelled landscape features in satellite data from London. We then incorporate landscape features from satellite image data from Pakistan, labelled using the CNN, in a well-known Susceptible-Infectious-Recovered epidemic model, alongside dengue case data from 2012-2016 in Pakistan. We study improvement of the prediction model for each of the individual landscape features, and assess the feasibility of using image labels from a different place. We find that incorporating satellite-derived landscape features can improve prediction of outbreaks, which is important for proactive and strategic surveillance and control programmes.
Population-aware Hierarchical Bayesian Domain Adaptation
Nov 21, 2018


Abstract:Population attributes are essential in health for understanding who the data represents and precision medicine efforts. Even within disease infection labels, patients can exhibit significant variability; "fever" may mean something different when reported in a doctor's office versus from an online app, precluding directly learning across different datasets for the same prediction task. This problem falls into the domain adaptation paradigm. However, research in this area has to-date not considered who generates the data; symptoms reported by a woman versus a man, for example, could also have different implications. We propose a novel population-aware domain adaptation approach by formulating the domain adaptation task as a multi-source hierarchical Bayesian framework. The model improves prediction in the case of largely unlabelled target data by harnessing both domain and population invariant information.
Domain Adaptation for Infection Prediction from Symptoms Based on Data from Different Study Designs and Contexts
Jun 22, 2018



Abstract:Acute respiratory infections have epidemic and pandemic potential and thus are being studied worldwide, albeit in many different contexts and study formats. Predicting infection from symptom data is critical, though using symptom data from varied studies in aggregate is challenging because the data is collected in different ways. Accordingly, different symptom profiles could be more predictive in certain studies, or even symptoms of the same name could have different meanings in different contexts. We assess state-of-the-art transfer learning methods for improving prediction of infection from symptom data in multiple types of health care data ranging from clinical, to home-visit as well as crowdsourced studies. We show interesting characteristics regarding six different study types and their feature domains. Further, we demonstrate that it is possible to use data collected from one study to predict infection in another, at close to or better than using a single dataset for prediction on itself. We also investigate in which conditions specific transfer learning and domain adaptation methods may perform better on symptom data. This work has the potential for broad applicability as we show how it is possible to transfer learning from one public health study design to another, and data collected from one study may be used for prediction of labels for another, even collected through different study designs, populations and contexts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge