Population-aware Hierarchical Bayesian Domain Adaptation via Multiple-component Invariant Learning
Paper and Code
Sep 13, 2019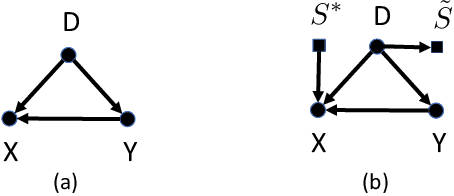
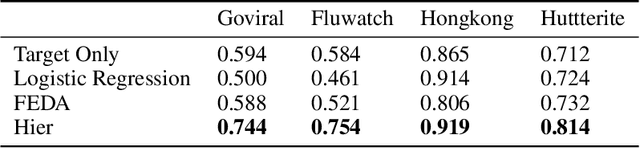
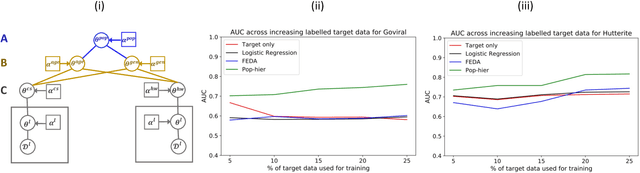
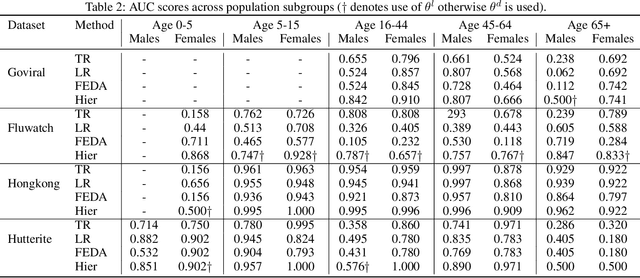
While machine learning is rapidly being developed and deployed in health settings such as influenza prediction, there are critical challenges in using data from one environment in another due to variability in features; even within disease labels there can be differences (e.g. "fever" may mean something different reported in a doctor's office versus in an online app). Moreover, models are often built on passive, observational data which contain different distributions of population subgroups (e.g. men or women). Thus, there are two forms of instability between environments in this observational transport problem. We first harness knowledge from health to conceptualize the underlying causal structure of this problem in a health outcome prediction task. Based on sources of stability in the model, we posit that for human-sourced data and health prediction tasks we can combine environment and population information in a novel population-aware hierarchical Bayesian domain adaptation framework that harnesses multiple invariant components through population attributes when needed. We study the conditions under which invariant learning fails, leading to reliance on the environment-specific attributes. Experimental results for an influenza prediction task on four datasets gathered from different contexts show the model can improve prediction in the case of largely unlabelled target data from a new environment and different constituent population, by harnessing both environment and population invariant information. This work represents a novel, principled way to address a critical challenge by blending domain (health) knowledge and algorithmic innovation. The proposed approach will have a significant impact in many social settings wherein who and where the data comes from matters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge