Mohamad Koohi-Moghadam
PathoGen: Diffusion-Based Synthesis of Realistic Lesions in Histopathology Images
Jan 13, 2026Abstract:The development of robust artificial intelligence models for histopathology diagnosis is severely constrained by the scarcity of expert-annotated lesion data, particularly for rare pathologies and underrepresented disease subtypes. While data augmentation offers a potential solution, existing methods fail to generate sufficiently realistic lesion morphologies that preserve the complex spatial relationships and cellular architectures characteristic of histopathological tissues. Here we present PathoGen, a diffusion-based generative model that enables controllable, high-fidelity inpainting of lesions into benign histopathology images. Unlike conventional augmentation techniques, PathoGen leverages the iterative refinement process of diffusion models to synthesize lesions with natural tissue boundaries, preserved cellular structures, and authentic staining characteristics. We validate PathoGen across four diverse datasets representing distinct diagnostic challenges: kidney, skin, breast, and prostate pathology. Quantitative assessment confirms that PathoGen outperforms state-of-the-art generative baselines, including conditional GAN and Stable Diffusion, in image fidelity and distributional similarity. Crucially, we show that augmenting training sets with PathoGen-synthesized lesions enhances downstream segmentation performance compared to traditional geometric augmentations, particularly in data-scarce regimes. Besides, by simultaneously generating realistic morphology and pixel-level ground truth, PathoGen effectively overcomes the manual annotation bottleneck. This approach offers a scalable pathway for developing generalizable medical AI systems despite limited expert-labeled data.
Enhancing Out-of-Distribution Detection in Medical Imaging with Normalizing Flows
Feb 17, 2025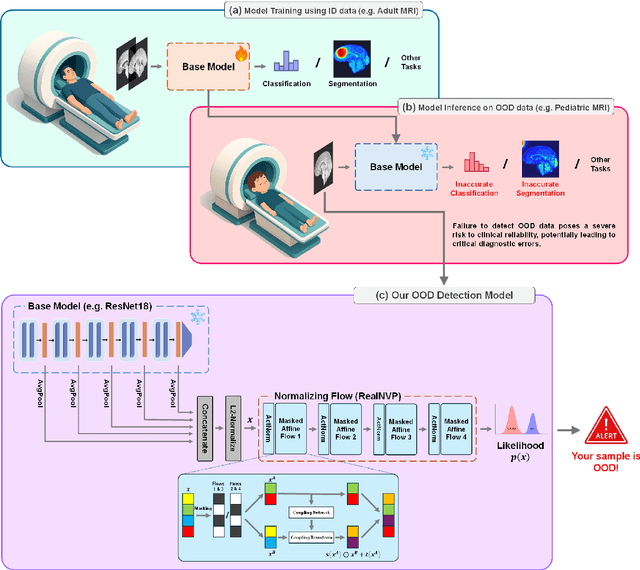
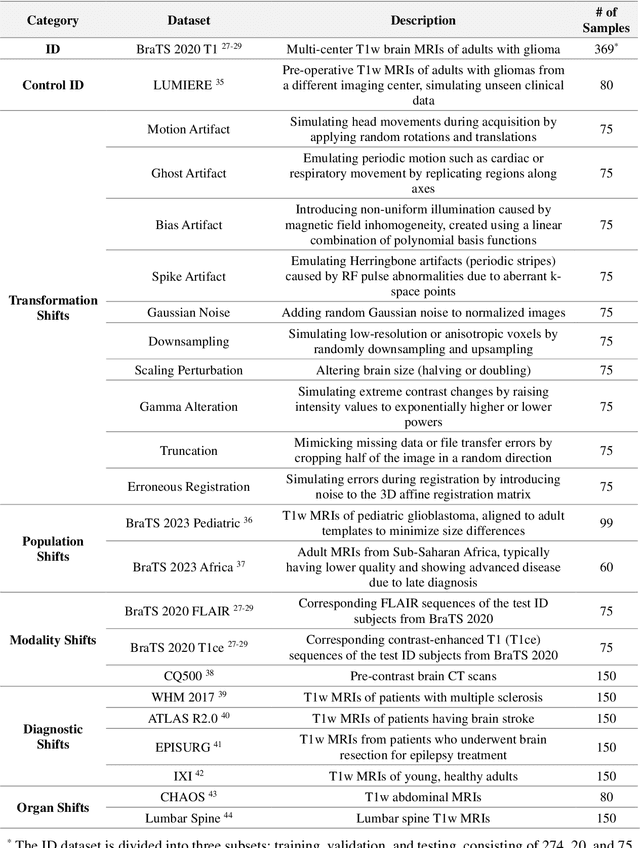
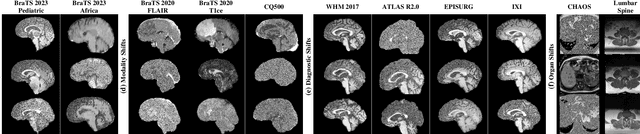
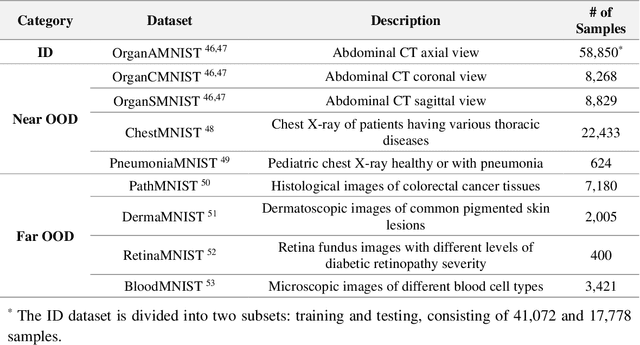
Abstract:Out-of-distribution (OOD) detection is crucial in AI-driven medical imaging to ensure reliability and safety by identifying inputs outside a model's training distribution. Existing methods often require retraining or modifications to pre-trained models, which is impractical for clinical applications. This study introduces a post-hoc normalizing flow-based approach that seamlessly integrates with pre-trained models. By leveraging normalizing flows, it estimates the likelihood of feature vectors extracted from pre-trained models, capturing semantically meaningful representations without relying on pixel-level statistics. The method was evaluated using the MedMNIST benchmark and a newly curated MedOOD dataset simulating clinically relevant distributional shifts. Performance was measured using standard OOD detection metrics (e.g., AUROC, FPR@95, AUPR_IN, AUPR_OUT), with statistical analyses comparing it against ten baseline methods. On MedMNIST, the proposed model achieved an AUROC of 93.80%, outperforming state-of-the-art methods. On MedOOD, it achieved an AUROC of 84.61%, demonstrating superior performance against other methods. Its post-hoc nature ensures compatibility with existing clinical workflows, addressing the limitations of previous approaches. The model and code to build OOD datasets are available at https://github.com/dlotfi/MedOODFlow.
Beta-Rank: A Robust Convolutional Filter Pruning Method For Imbalanced Medical Image Analysis
Apr 15, 2023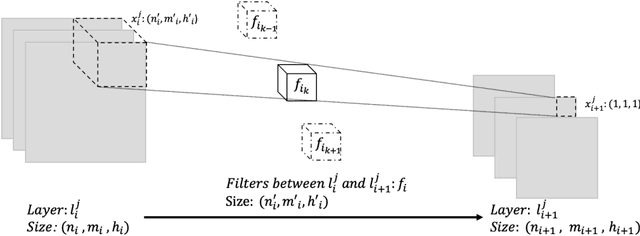
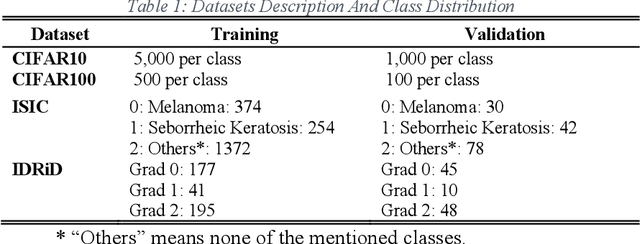
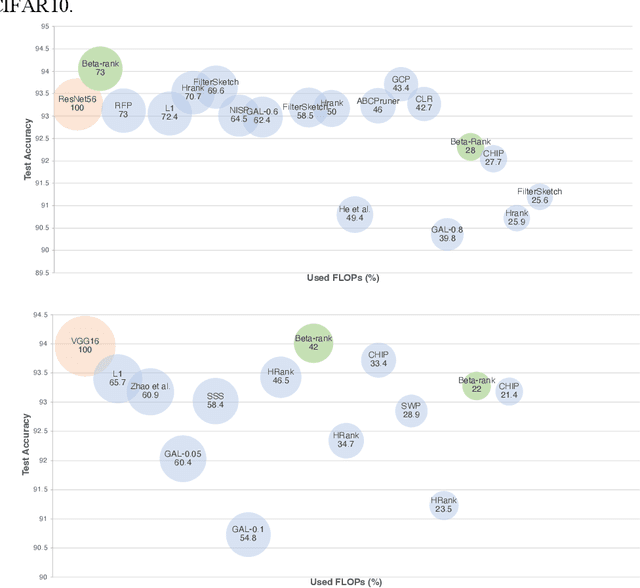
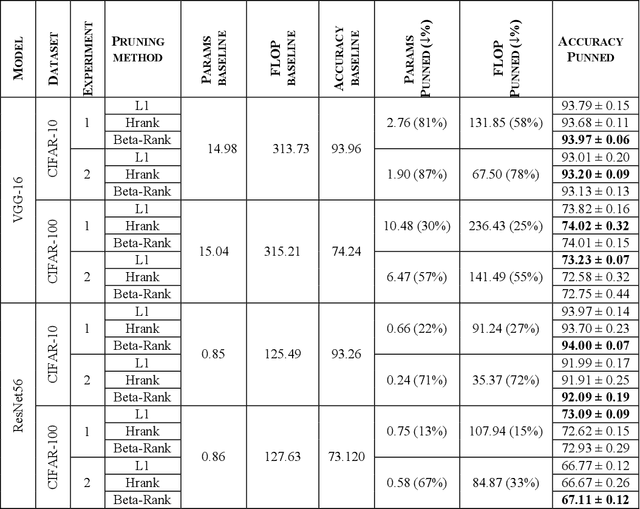
Abstract:As deep neural networks include a high number of parameters and operations, it can be a challenge to implement these models on devices with limited computational resources. Despite the development of novel pruning methods toward resource-efficient models, it has become evident that these models are not capable of handling "imbalanced" and "limited number of data points". With input and output information, along with the values of the filters, a novel filter pruning method is proposed. Our pruning method considers the fact that all information about the importance of a filter may not be reflected in the value of the filter. Instead, it is reflected in the changes made to the data after the filter is applied to it. In this work, three methods are compared with the same training conditions except for the ranking of each method. We demonstrated that our model performed significantly better than other methods for medical datasets which are inherently imbalanced. When we removed up to 58% of FLOPs for the IDRID dataset and up to 45% for the ISIC dataset, our model was able to yield an equivalent (or even superior) result to the baseline model while other models were unable to achieve similar results. To evaluate FLOP and parameter reduction using our model in real-world settings, we built a smartphone app, where we demonstrated a reduction of up to 79% in memory usage and 72% in prediction time. All codes and parameters for training different models are available at https://github.com/mohofar/Beta-Rank
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge