Ming-Chen Hsu
A Physics-Guided Neural Operator Learning Approach to Model Biological Tissues from Digital Image Correlation Measurements
Apr 01, 2022
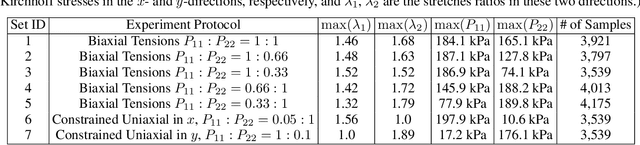
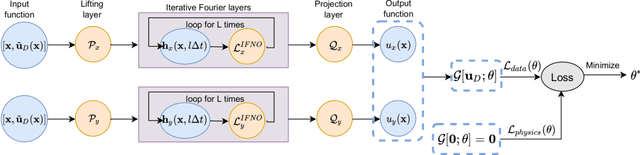
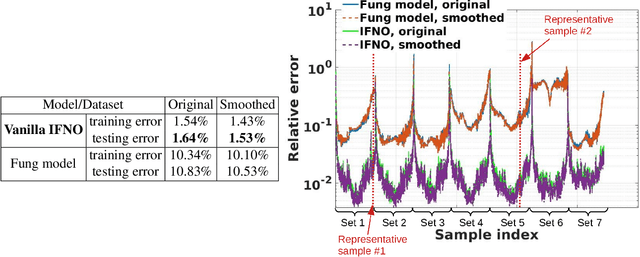
Abstract:We present a data-driven workflow to biological tissue modeling, which aims to predict the displacement field based on digital image correlation (DIC) measurements under unseen loading scenarios, without postulating a specific constitutive model form nor possessing knowledges on the material microstructure. To this end, a material database is constructed from the DIC displacement tracking measurements of multiple biaxial stretching protocols on a porcine tricuspid valve anterior leaflet, with which we build a neural operator learning model. The material response is modeled as a solution operator from the loading to the resultant displacement field, with the material microstructure properties learned implicitly from the data and naturally embedded in the network parameters. Using various combinations of loading protocols, we compare the predictivity of this framework with finite element analysis based on the phenomenological Fung-type model. From in-distribution tests, the predictivity of our approach presents good generalizability to different loading conditions and outperforms the conventional constitutive modeling at approximately one order of magnitude. When tested on out-of-distribution loading ratios, the neural operator learning approach becomes less effective. To improve the generalizability of our framework, we propose a physics-guided neural operator learning model via imposing partial physics knowledge. This method is shown to improve the model's extrapolative performance in the small-deformation regime. Our results demonstrate that with sufficient data coverage and/or guidance from partial physics constraints, the data-driven approach can be a more effective method for modeling biological materials than the traditional constitutive modeling.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge