Michael T. C. Ying
Decompose-and-Integrate Learning for Multi-class Segmentation in Medical Images
Jun 07, 2019



Abstract:Segmentation maps of medical images annotated by medical experts contain rich spatial information. In this paper, we propose to decompose annotation maps to learn disentangled and richer feature transforms for segmentation problems in medical images. Our new scheme consists of two main stages: decompose and integrate. Decompose: by annotation map decomposition, the original segmentation problem is decomposed into multiple segmentation sub-problems; these new segmentation sub-problems are modeled by training multiple deep learning modules, each with its own set of feature transforms. Integrate: a procedure summarizes the solutions of the modules in the previous stage; a final solution is then formed for the original segmentation problem. Multiple ways of annotation map decomposition are presented and a new end-to-end trainable K-to-1 deep network framework is developed for implementing our proposed "decompose-and-integrate" learning scheme. In experiments, we demonstrate that our decompose-and-integrate segmentation, utilizing state-of-the-art fully convolutional networks (e.g., DenseVoxNet in 3D and CUMedNet in 2D), improves segmentation performance on multiple 3D and 2D datasets. Ablation study confirms the effectiveness of our proposed learning scheme for medical images.
BoxNet: Deep Learning Based Biomedical Image Segmentation Using Boxes Only Annotation
Jun 02, 2018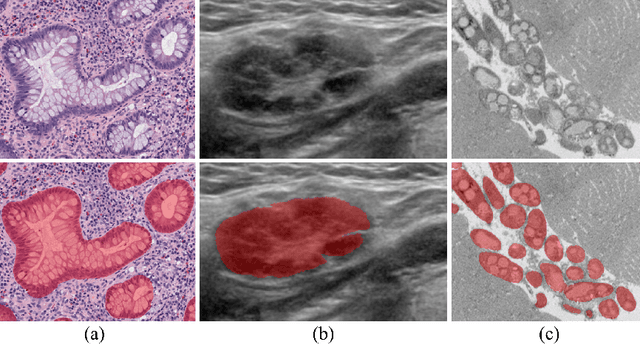



Abstract:In recent years, deep learning (DL) methods have become powerful tools for biomedical image segmentation. However, high annotation efforts and costs are commonly needed to acquire sufficient biomedical training data for DL models. To alleviate the burden of manual annotation, in this paper, we propose a new weakly supervised DL approach for biomedical image segmentation using boxes only annotation. First, we develop a method to combine graph search (GS) and DL to generate fine object masks from box annotation, in which DL uses box annotation to compute a rough segmentation for GS and then GS is applied to locate the optimal object boundaries. During the mask generation process, we carefully utilize information from box annotation to filter out potential errors, and then use the generated masks to train an accurate DL segmentation network. Extensive experiments on gland segmentation in histology images, lymph node segmentation in ultrasound images, and fungus segmentation in electron microscopy images show that our approach attains superior performance over the best known state-of-the-art weakly supervised DL method and is able to achieve (1) nearly the same accuracy compared to fully supervised DL methods with far less annotation effort, (2) significantly better results with similar annotation time, and (3) robust performance in various applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge