Michael Bonham
Using Whole Slide Image Representations from Self-Supervised Contrastive Learning for Melanoma Concordance Regression
Oct 10, 2022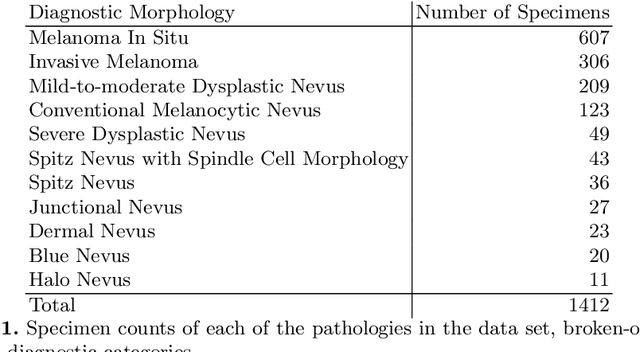
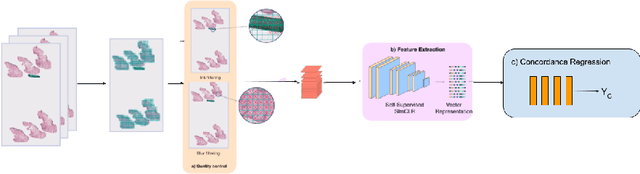
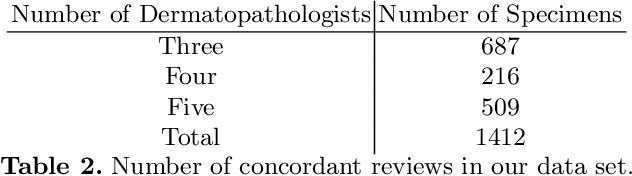
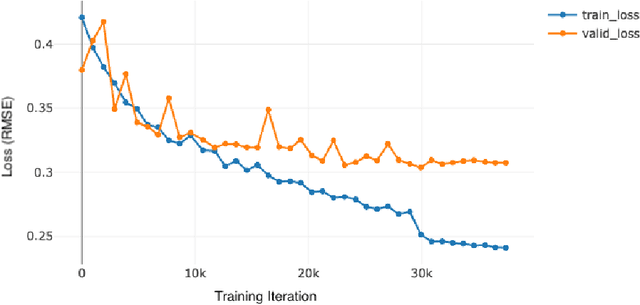
Abstract:Although melanoma occurs more rarely than several other skin cancers, patients' long term survival rate is extremely low if the diagnosis is missed. Diagnosis is complicated by a high discordance rate among pathologists when distinguishing between melanoma and benign melanocytic lesions. A tool that provides potential concordance information to healthcare providers could help inform diagnostic, prognostic, and therapeutic decision-making for challenging melanoma cases. We present a melanoma concordance regression deep learning model capable of predicting the concordance rate of invasive melanoma or melanoma in-situ from digitized Whole Slide Images (WSIs). The salient features corresponding to melanoma concordance were learned in a self-supervised manner with the contrastive learning method, SimCLR. We trained a SimCLR feature extractor with 83,356 WSI tiles randomly sampled from 10,895 specimens originating from four distinct pathology labs. We trained a separate melanoma concordance regression model on 990 specimens with available concordance ground truth annotations from three pathology labs and tested the model on 211 specimens. We achieved a Root Mean Squared Error (RMSE) of 0.28 +/- 0.01 on the test set. We also investigated the performance of using the predicted concordance rate as a malignancy classifier, and achieved a precision and recall of 0.85 +/- 0.05 and 0.61 +/- 0.06, respectively, on the test set. These results are an important first step for building an artificial intelligence (AI) system capable of predicting the results of consulting a panel of experts and delivering a score based on the degree to which the experts would agree on a particular diagnosis. Such a system could be used to suggest additional testing or other action such as ordering additional stains or genetic tests.
A Pathology Deep Learning System Capable of Triage of Melanoma Specimens Utilizing Dermatopathologist Consensus as Ground Truth
Sep 15, 2021



Abstract:Although melanoma occurs more rarely than several other skin cancers, patients' long term survival rate is extremely low if the diagnosis is missed. Diagnosis is complicated by a high discordance rate among pathologists when distinguishing between melanoma and benign melanocytic lesions. A tool that allows pathology labs to sort and prioritize melanoma cases in their workflow could improve turnaround time by prioritizing challenging cases and routing them directly to the appropriate subspecialist. We present a pathology deep learning system (PDLS) that performs hierarchical classification of digitized whole slide image (WSI) specimens into six classes defined by their morphological characteristics, including classification of "Melanocytic Suspect" specimens likely representing melanoma or severe dysplastic nevi. We trained the system on 7,685 images from a single lab (the reference lab), including the the largest set of triple-concordant melanocytic specimens compiled to date, and tested the system on 5,099 images from two distinct validation labs. We achieved Area Underneath the ROC Curve (AUC) values of 0.93 classifying Melanocytic Suspect specimens on the reference lab, 0.95 on the first validation lab, and 0.82 on the second validation lab. We demonstrate that the PDLS is capable of automatically sorting and triaging skin specimens with high sensitivity to Melanocytic Suspect cases and that a pathologist would only need between 30% and 60% of the caseload to address all melanoma specimens.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge