Melanie Rey
University of Basel
DiscoGen: Learning to Discover Gene Regulatory Networks
Apr 12, 2023Abstract:Accurately inferring Gene Regulatory Networks (GRNs) is a critical and challenging task in biology. GRNs model the activatory and inhibitory interactions between genes and are inherently causal in nature. To accurately identify GRNs, perturbational data is required. However, most GRN discovery methods only operate on observational data. Recent advances in neural network-based causal discovery methods have significantly improved causal discovery, including handling interventional data, improvements in performance and scalability. However, applying state-of-the-art (SOTA) causal discovery methods in biology poses challenges, such as noisy data and a large number of samples. Thus, adapting the causal discovery methods is necessary to handle these challenges. In this paper, we introduce DiscoGen, a neural network-based GRN discovery method that can denoise gene expression measurements and handle interventional data. We demonstrate that our model outperforms SOTA neural network-based causal discovery methods.
Copula Mixture Model for Dependency-seeking Clustering
Jun 27, 2012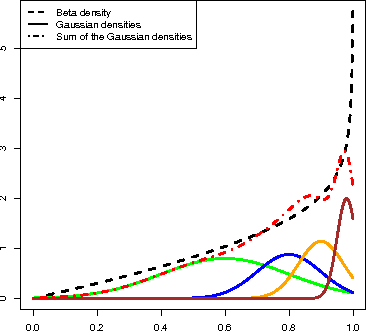
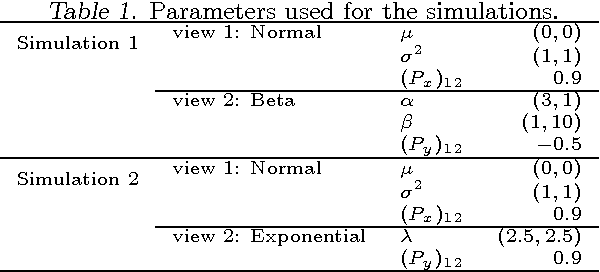
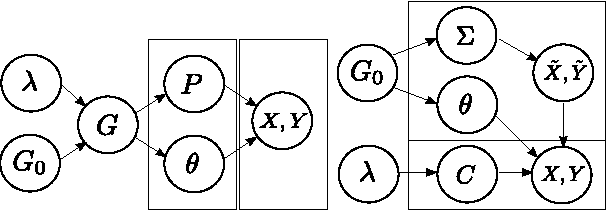
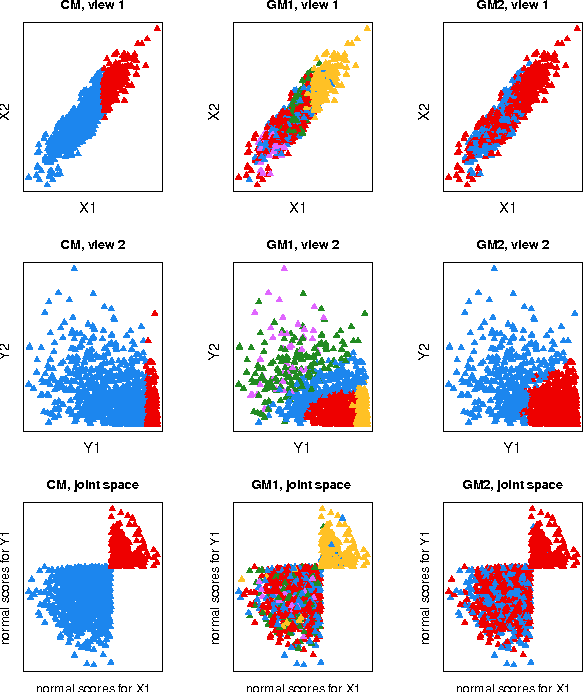
Abstract:We introduce a copula mixture model to perform dependency-seeking clustering when co-occurring samples from different data sources are available. The model takes advantage of the great flexibility offered by the copulas framework to extend mixtures of Canonical Correlation Analysis to multivariate data with arbitrary continuous marginal densities. We formulate our model as a non-parametric Bayesian mixture, while providing efficient MCMC inference. Experiments on synthetic and real data demonstrate that the increased flexibility of the copula mixture significantly improves the clustering and the interpretability of the results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge