Meir Perez
DLT: Conditioned layout generation with Joint Discrete-Continuous Diffusion Layout Transformer
Mar 07, 2023Abstract:Generating visual layouts is an essential ingredient of graphic design. The ability to condition layout generation on a partial subset of component attributes is critical to real-world applications that involve user interaction. Recently, diffusion models have demonstrated high-quality generative performances in various domains. However, it is unclear how to apply diffusion models to the natural representation of layouts which consists of a mix of discrete (class) and continuous (location, size) attributes. To address the conditioning layout generation problem, we introduce DLT, a joint discrete-continuous diffusion model. DLT is a transformer-based model which has a flexible conditioning mechanism that allows for conditioning on any given subset of all the layout component classes, locations, and sizes. Our method outperforms state-of-the-art generative models on various layout generation datasets with respect to different metrics and conditioning settings. Additionally, we validate the effectiveness of our proposed conditioning mechanism and the joint continuous-diffusion process. This joint process can be incorporated into a wide range of mixed discrete-continuous generative tasks.
The fuzzy gene filter: A classifier performance assesment
Aug 23, 2011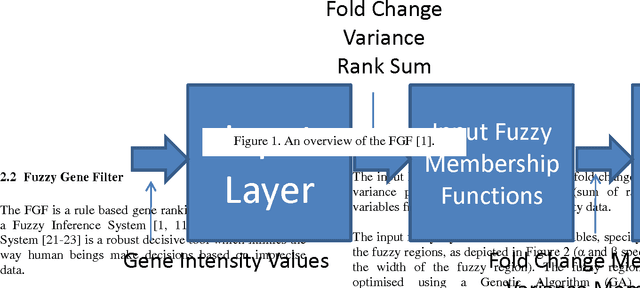
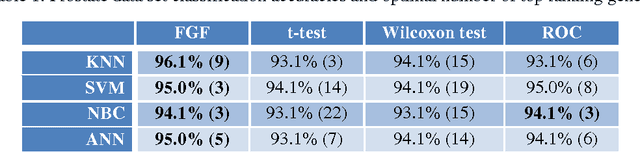
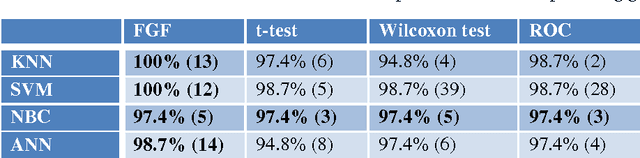
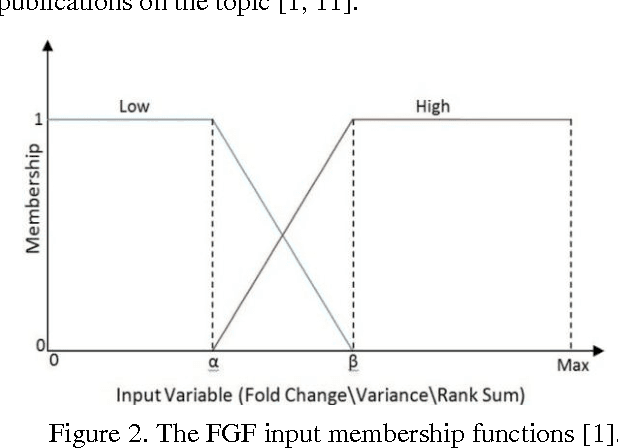
Abstract:The Fuzzy Gene Filter (FGF) is an optimised Fuzzy Inference System designed to rank genes in order of differential expression, based on expression data generated in a microarray experiment. This paper examines the effectiveness of the FGF for feature selection using various classification architectures. The FGF is compared to three of the most common gene ranking algorithms: t-test, Wilcoxon test and ROC curve analysis. Four classification schemes are used to compare the performance of the FGF vis-a-vis the standard approaches: K Nearest Neighbour (KNN), Support Vector Machine (SVM), Naive Bayesian Classifier (NBC) and Artificial Neural Network (ANN). A nested stratified Leave-One-Out Cross Validation scheme is used to identify the optimal number top ranking genes, as well as the optimal classifier parameters. Two microarray data sets are used for the comparison: a prostate cancer data set and a lymphoma data set.
Stochastic Optimization Approaches for Solving Sudoku
May 06, 2008



Abstract:In this paper the Sudoku problem is solved using stochastic search techniques and these are: Cultural Genetic Algorithm (CGA), Repulsive Particle Swarm Optimization (RPSO), Quantum Simulated Annealing (QSA) and the Hybrid method that combines Genetic Algorithm with Simulated Annealing (HGASA). The results obtained show that the CGA, QSA and HGASA are able to solve the Sudoku puzzle with CGA finding a solution in 28 seconds, while QSA finding a solution in 65 seconds and HGASA in 1.447 seconds. This is mainly because HGASA combines the parallel searching of GA with the flexibility of SA. The RPSO was found to be unable to solve the puzzle.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge