Mehul S Raval
Person Retrieval in Surveillance Using Textual Query: A Review
May 06, 2021



Abstract:Recent advancement of research in biometrics, computer vision, and natural language processing has discovered opportunities for person retrieval from surveillance videos using textual query. The prime objective of a surveillance system is to locate a person using a description, e.g., a short woman with a pink t-shirt and white skirt carrying a black purse. She has brown hair. Such a description contains attributes like gender, height, type of clothing, colour of clothing, hair colour, and accessories. Such attributes are formally known as soft biometrics. They help bridge the semantic gap between a human description and a machine as a textual query contains the person's soft biometric attributes. It is also not feasible to manually search through huge volumes of surveillance footage to retrieve a specific person. Hence, automatic person retrieval using vision and language-based algorithms is becoming popular. In comparison to other state-of-the-art reviews, the contribution of the paper is as follows: 1. Recommends most discriminative soft biometrics for specifiic challenging conditions. 2. Integrates benchmark datasets and retrieval methods for objective performance evaluation. 3. A complete snapshot of techniques based on features, classifiers, number of soft biometric attributes, type of the deep neural networks, and performance measures. 4. The comprehensive coverage of person retrieval from handcrafted features based methods to end-to-end approaches based on natural language description.
* 45 pages, 17 figures, 6 Tables
A Survey and Analysis on Automated Glioma Brain Tumor Segmentation and Overall Patient Survival Prediction
Jan 26, 2021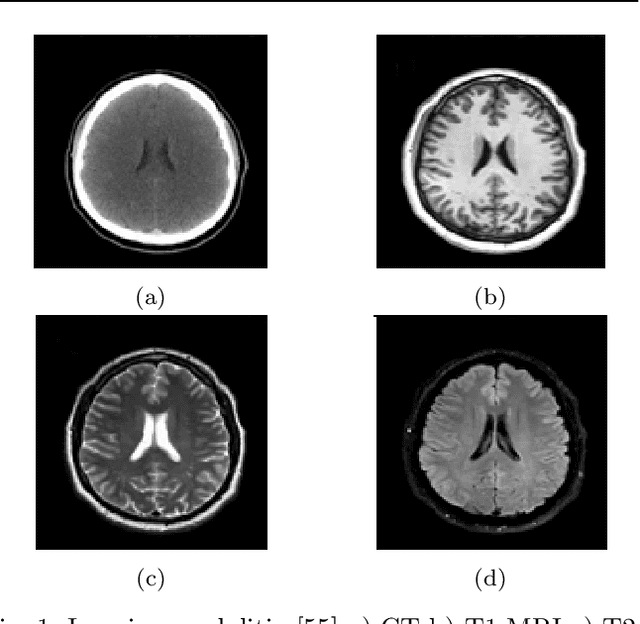
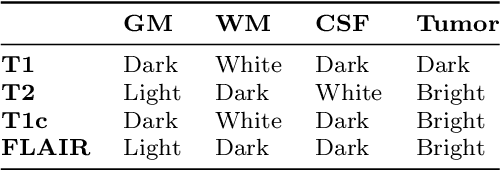
Abstract:Glioma is the most deadly brain tumor with high mortality. Treatment planning by human experts depends on the proper diagnosis of physical symptoms along with Magnetic Resonance(MR) image analysis. Highly variability of a brain tumor in terms of size, shape, location, and a high volume of MR images makes the analysis time-consuming. Automatic segmentation methods achieve a reduction in time with excellent reproducible results. The article aims to survey the advancement of automated methods for Glioma brain tumor segmentation. It is also essential to make an objective evaluation of various models based on the benchmark. Therefore, the 2012 - 2019 BraTS challenges database evaluates state-of-the-art methods. The complexity of tasks under the challenge has grown from segmentation (Task1) to overall survival prediction (Task 2) to uncertainty prediction for classification (Task 3). The paper covers the complete gamut of brain tumor segmentation using handcrafted features to deep neural network models for Task 1. The aim is to showcase a complete change of trends in automated brain tumor models. The paper also covers end to end joint models involving brain tumor segmentation and overall survival prediction. All the methods are probed, and parameters that affect performance are tabulated and analyzed.
* 40 pages, 19 figures, 11 Tables
Glioblastoma Multiforme Patient Survival Prediction
Jan 26, 2021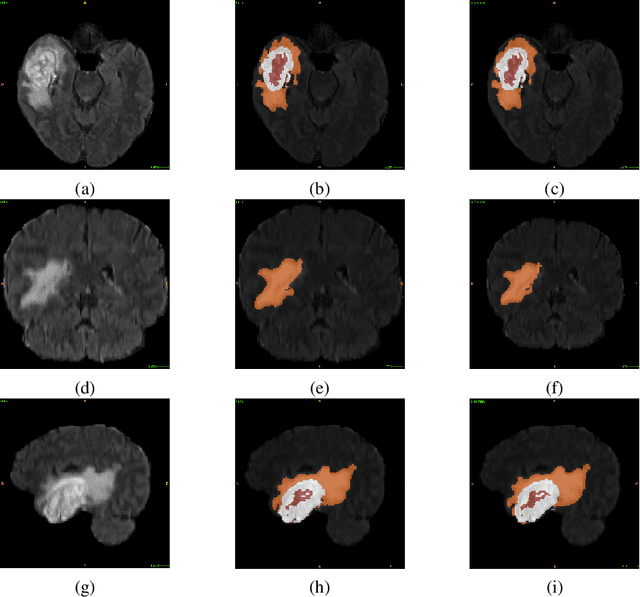
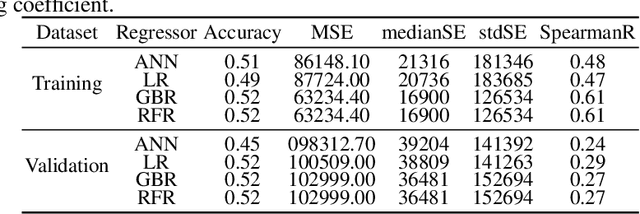

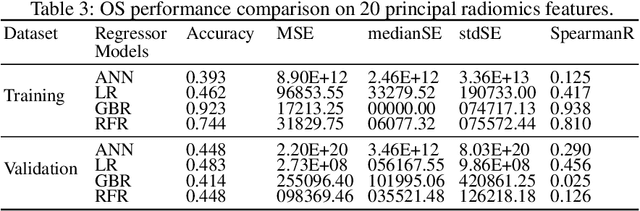
Abstract:Glioblastoma Multiforme is a very aggressive type of brain tumor. Due to spatial and temporal intra-tissue inhomogeneity, location and the extent of the cancer tissue, it is difficult to detect and dissect the tumor regions. In this paper, we propose survival prognosis models using four regressors operating on handcrafted image-based and radiomics features. We hypothesize that the radiomics shape features have the highest correlation with survival prediction. The proposed approaches were assessed on the Brain Tumor Segmentation (BraTS-2020) challenge dataset. The highest accuracy of image features with random forest regressor approach was 51.5\% for the training and 51.7\% for the validation dataset. The gradient boosting regressor with shape features gave an accuracy of 91.5\% and 62.1\% on training and validation datasets respectively. It is better than the BraTS 2020 survival prediction challenge winners on the training and validation datasets. Our work shows that handcrafted features exhibit a strong correlation with survival prediction. The consensus based regressor with gradient boosting and radiomics shape features is the best combination for survival prediction.
* 10 pages, 9 figures
3D Semantic Segmentation of Brain Tumor for Overall Survival Prediction
Aug 25, 2020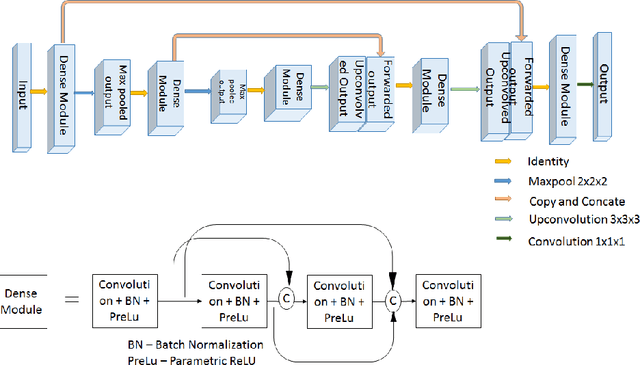
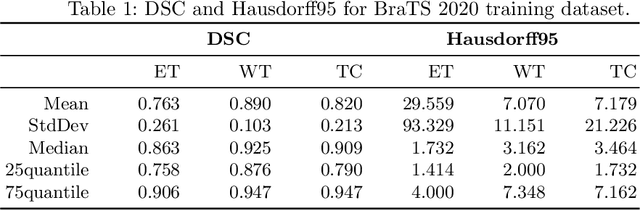
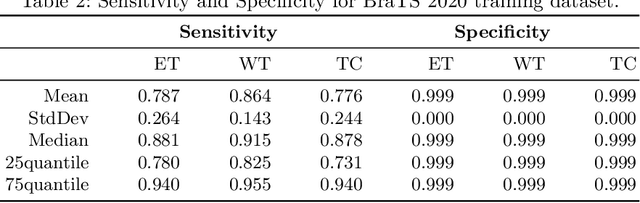
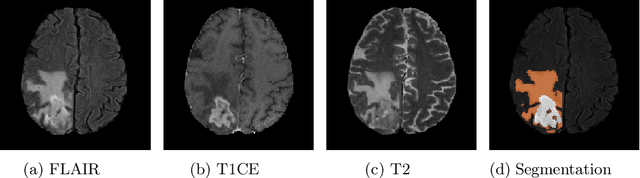
Abstract:Glioma, the malignant brain tumor, requires immediate treatment to improve the survival of patients. Gliomas heterogeneous nature makes the segmentation difficult, especially for sub-regions like necrosis, enhancing tumor, non-enhancing tumor, and Edema. Deep neural networks like full convolution neural networks and ensemble of fully convolution neural networks are successful for Glioma segmentation. The paper demonstrates the use of a 3D fully convolution neural network with a three layer encoder decoder approach for layer arrangement. The encoder blocks include the dense modules, and decoder blocks include convolution modules. The input to the network is 3D patches. The loss function combines dice loss and focal loss functions. The validation set dice score of the network is 0.74, 0.88, and 0.73 for enhancing tumor, whole tumor, and tumor core, respectively. The Random Forest Regressor uses shape, volumetric, and age features extracted from ground truth for overall survival prediction. The regressor achieves an accuracy of 44.8% on the validation set.
A Review on End-To-End Methods for Brain Tumor Segmentation and Overall Survival Prediction
May 31, 2020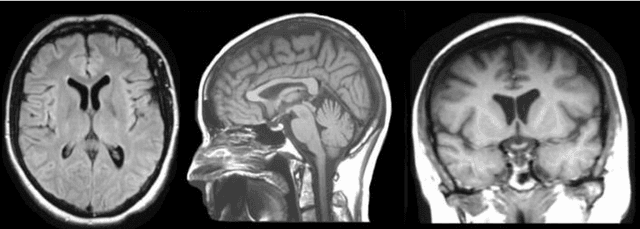
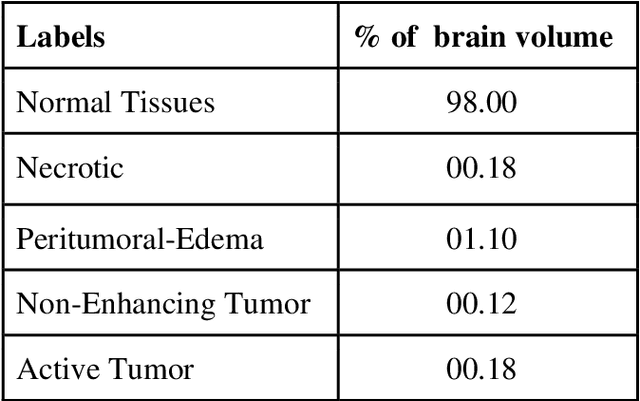
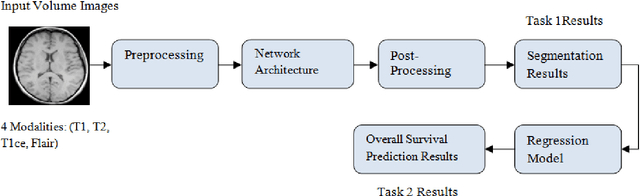
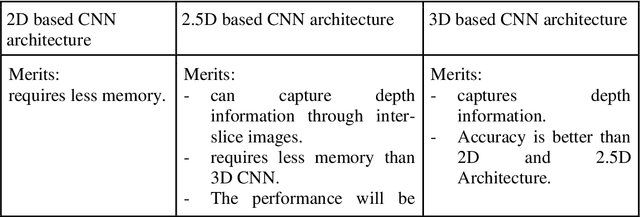
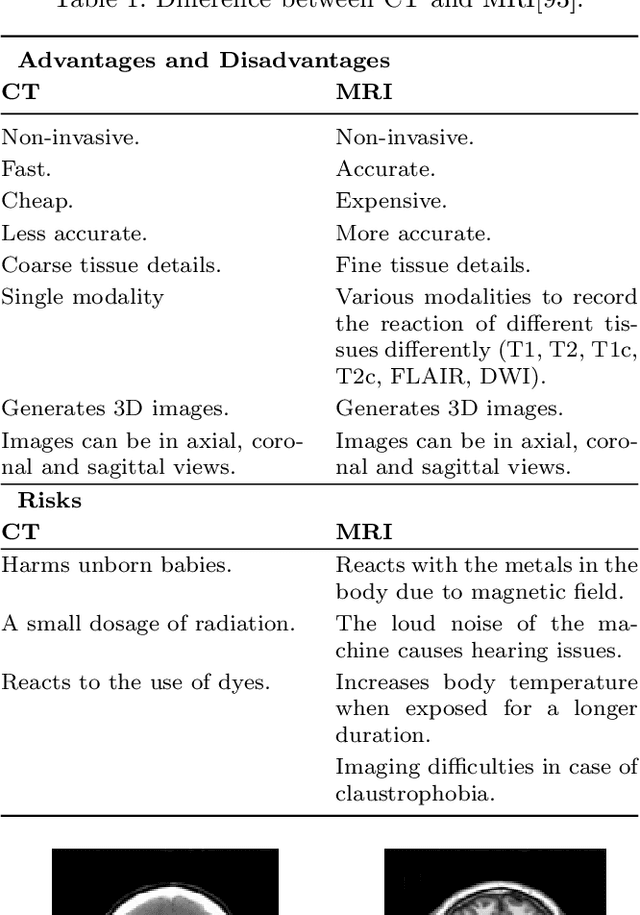
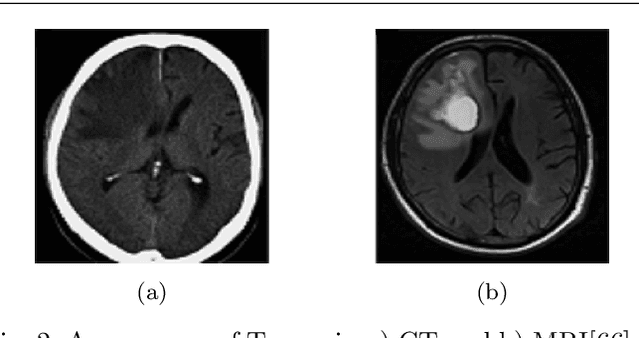
Abstract:Brain tumor segmentation intends to delineate tumor tissues from healthy brain tissues. The tumor tissues include necrosis, peritumoral edema, and active tumor. In contrast, healthy brain tissues include white matter, gray matter, and cerebrospinal fluid. The MRI based brain tumor segmentation research is gaining popularity as; 1. It does not irradiate ionized radiation like X-ray or computed tomography imaging. 2. It produces detailed pictures of internal body structures. The MRI scans are input to deep learning-based approaches which are useful for automatic brain tumor segmentation. The features from segments are fed to the classifier which predict the overall survival of the patient. The motive of this paper is to give an extensive overview of state-of-the-art jointly covering brain tumor segmentation and overall survival prediction.
Visual Appearance Based Person Retrieval in Unconstrained Environment Videos
Oct 31, 2019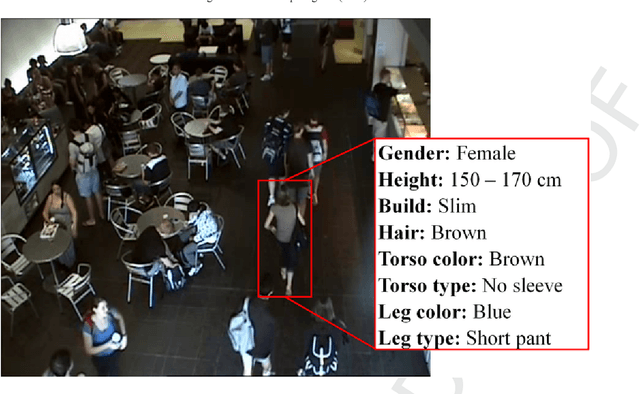
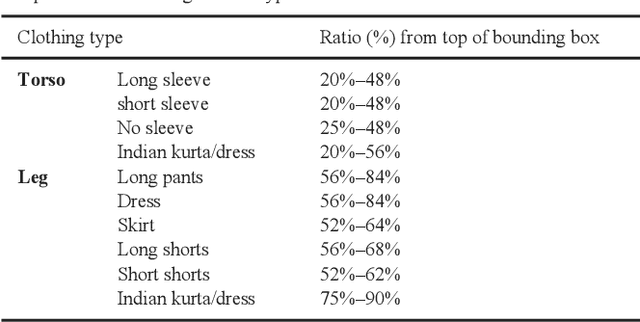
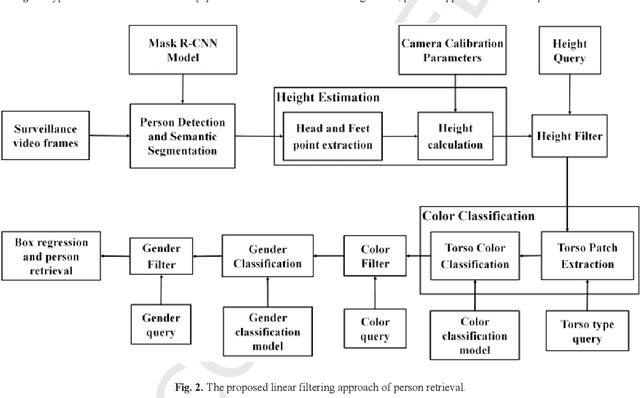

Abstract:Visual appearance-based person retrieval is a challenging problem in surveillance. It uses attributes like height, cloth color, cloth type and gender to describe a human. Such attributes are known as soft biometrics. This paper proposes person retrieval from surveillance video using height, torso cloth type, torso cloth color and gender. The approach introduces an adaptive torso patch extraction and bounding box regression to improve the retrieval. The algorithm uses fine-tuned Mask R-CNN and DenseNet-169 for person detection and attribute classification respectively. The performance is analyzed on AVSS 2018 challenge II dataset and it achieves 11.35% improvement over state-of-the-art based on average Intersection over Union measure.
* 11 pages
Brain Tumor Segmentation and Survival Prediction
Sep 20, 2019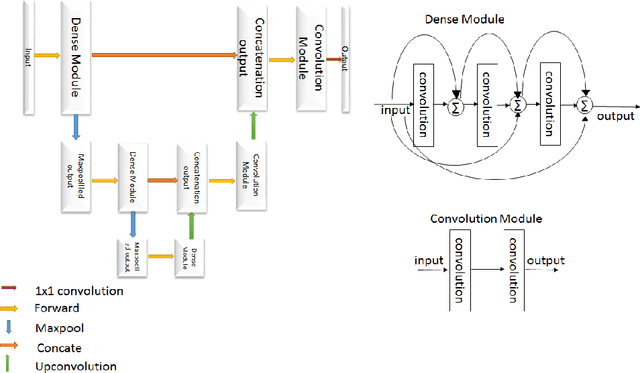
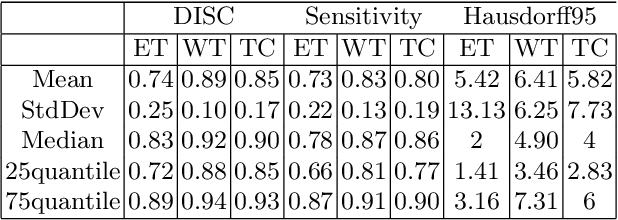
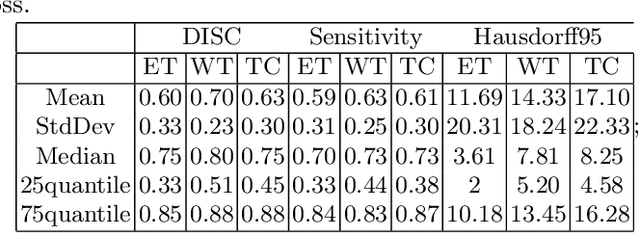
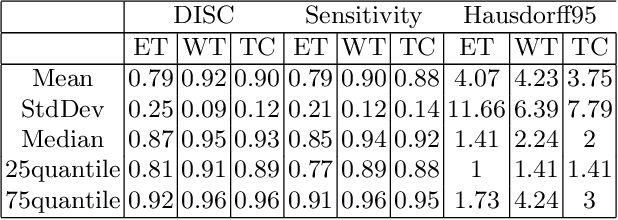
Abstract:The paper demonstrates the use of the fully convolutional neural network for glioma segmentation on the BraTS 2019 dataset. Three-layers deep encoder-decoder architecture is used along with dense connection at encoder part to propagate the information from coarse layer to deep layers. This architecture is used to train three tumor sub-components separately. Subcomponent training weights are initialized with whole tumor weights to get the localization of the tumor within the brain. At the end, three segmentation results were merged to get the entire tumor segmentation. Dice Similarity of training dataset with focal loss implementation for whole tumor, tumor core and enhancing tumor is 0.92, 0.90 and 0.79 respectively. Radiomic features along with segmentation results and age are used to predict the overall survival of patients using random forest regressor to classify survival of patients in long, medium and short survival classes. 55.4% of classification accuracy is reported for training dataset with the scans whose resection status is gross-total resection.
* 9 Pages
Prediction of Overall Survival of Brain Tumor Patients
Sep 10, 2019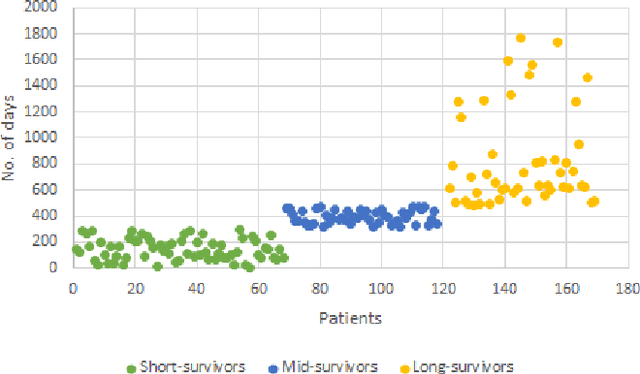
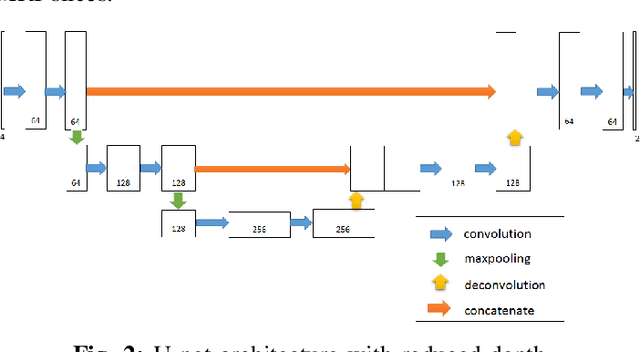
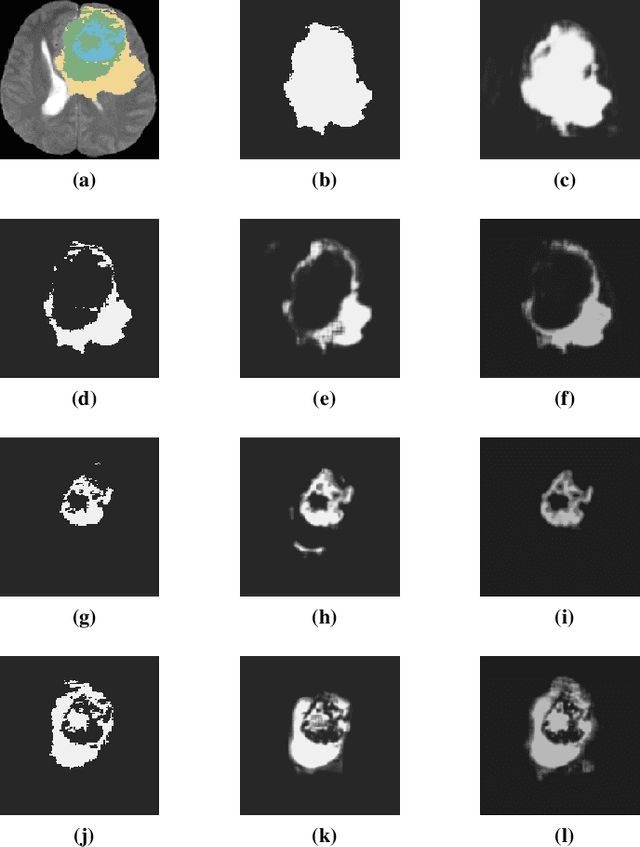
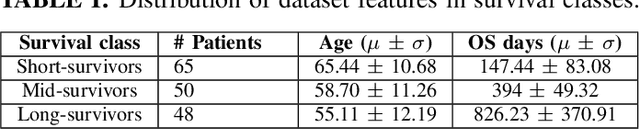
Abstract:Automated brain tumor segmentation plays an important role in the diagnosis and prognosis of the patient. In addition, features from the tumorous brain help in predicting patients overall survival. The main focus of this paper is to segment tumor from BRATS 2018 benchmark dataset and use age, shape and volumetric features to predict overall survival of patients. The random forest classifier achieves overall survival accuracy of 59% on the test dataset and 67% on the dataset with resection status as gross total resection. The proposed approach uses fewer features but achieves better accuracy than state of the art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge