Snehal Rajput
Glioblastoma Multiforme Patient Survival Prediction
Jan 26, 2021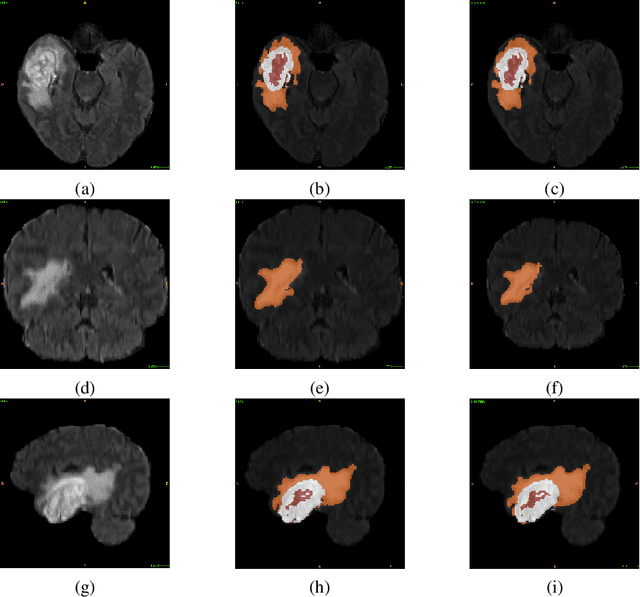
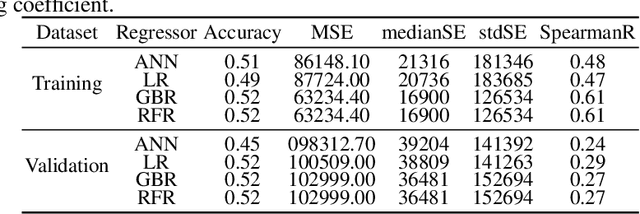

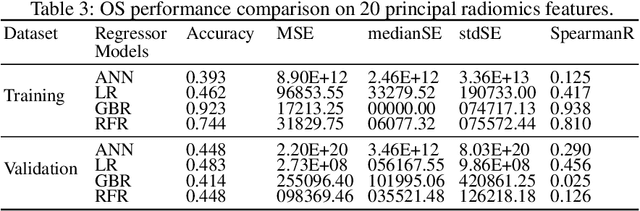
Abstract:Glioblastoma Multiforme is a very aggressive type of brain tumor. Due to spatial and temporal intra-tissue inhomogeneity, location and the extent of the cancer tissue, it is difficult to detect and dissect the tumor regions. In this paper, we propose survival prognosis models using four regressors operating on handcrafted image-based and radiomics features. We hypothesize that the radiomics shape features have the highest correlation with survival prediction. The proposed approaches were assessed on the Brain Tumor Segmentation (BraTS-2020) challenge dataset. The highest accuracy of image features with random forest regressor approach was 51.5\% for the training and 51.7\% for the validation dataset. The gradient boosting regressor with shape features gave an accuracy of 91.5\% and 62.1\% on training and validation datasets respectively. It is better than the BraTS 2020 survival prediction challenge winners on the training and validation datasets. Our work shows that handcrafted features exhibit a strong correlation with survival prediction. The consensus based regressor with gradient boosting and radiomics shape features is the best combination for survival prediction.
* 10 pages, 9 figures
A Review on End-To-End Methods for Brain Tumor Segmentation and Overall Survival Prediction
May 31, 2020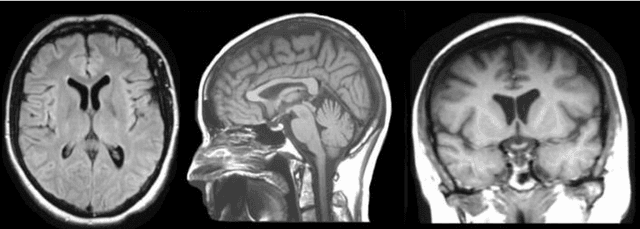
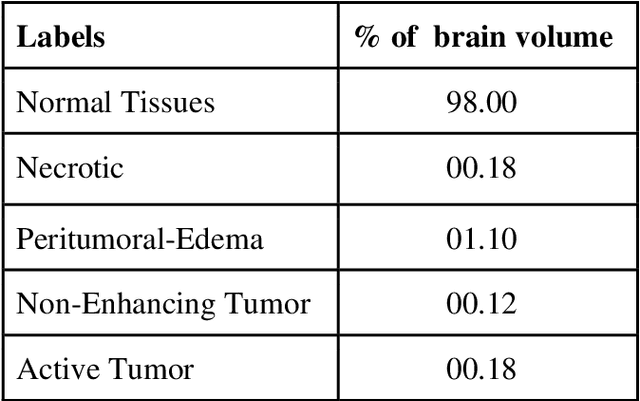
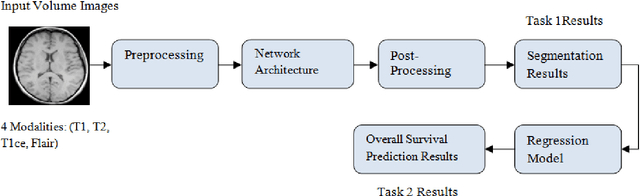
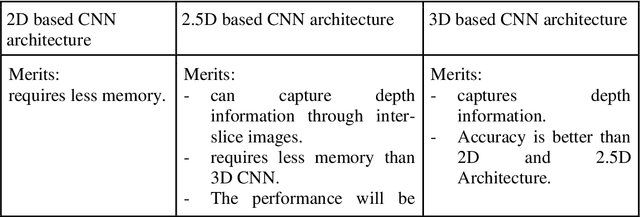
Abstract:Brain tumor segmentation intends to delineate tumor tissues from healthy brain tissues. The tumor tissues include necrosis, peritumoral edema, and active tumor. In contrast, healthy brain tissues include white matter, gray matter, and cerebrospinal fluid. The MRI based brain tumor segmentation research is gaining popularity as; 1. It does not irradiate ionized radiation like X-ray or computed tomography imaging. 2. It produces detailed pictures of internal body structures. The MRI scans are input to deep learning-based approaches which are useful for automatic brain tumor segmentation. The features from segments are fed to the classifier which predict the overall survival of the patient. The motive of this paper is to give an extensive overview of state-of-the-art jointly covering brain tumor segmentation and overall survival prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge