Md Hafizur Rahman
How a Bit Becomes a Story: Semantic Steering via Differentiable Fault Injection
Dec 09, 2025Abstract:Hard-to-detect hardware bit flips, from either malicious circuitry or bugs, have already been shown to make transformers vulnerable in non-generative tasks. This work, for the first time, investigates how low-level, bitwise perturbations (fault injection) to the weights of a large language model (LLM) used for image captioning can influence the semantic meaning of its generated descriptions while preserving grammatical structure. While prior fault analysis methods have shown that flipping a few bits can crash classifiers or degrade accuracy, these approaches overlook the semantic and linguistic dimensions of generative systems. In image captioning models, a single flipped bit might subtly alter how visual features map to words, shifting the entire narrative an AI tells about the world. We hypothesize that such semantic drifts are not random but differentiably estimable. That is, the model's own gradients can predict which bits, if perturbed, will most strongly influence meaning while leaving syntax and fluency intact. We design a differentiable fault analysis framework, BLADE (Bit-level Fault Analysis via Differentiable Estimation), that uses gradient-based sensitivity estimation to locate semantically critical bits and then refines their selection through a caption-level semantic-fluency objective. Our goal is not merely to corrupt captions, but to understand how meaning itself is encoded, distributed, and alterable at the bit level, revealing that even imperceptible low-level changes can steer the high-level semantics of generative vision-language models. It also opens pathways for robustness testing, adversarial defense, and explainable AI, by exposing how structured bit-level faults can reshape a model's semantic output.
ILASH: A Predictive Neural Architecture Search Framework for Multi-Task Applications
Dec 03, 2024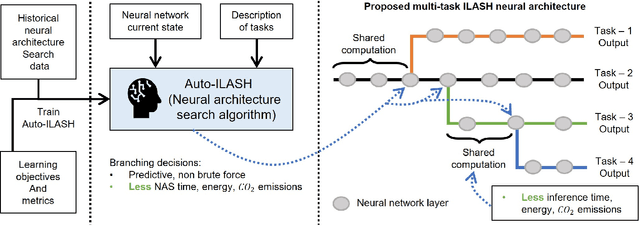
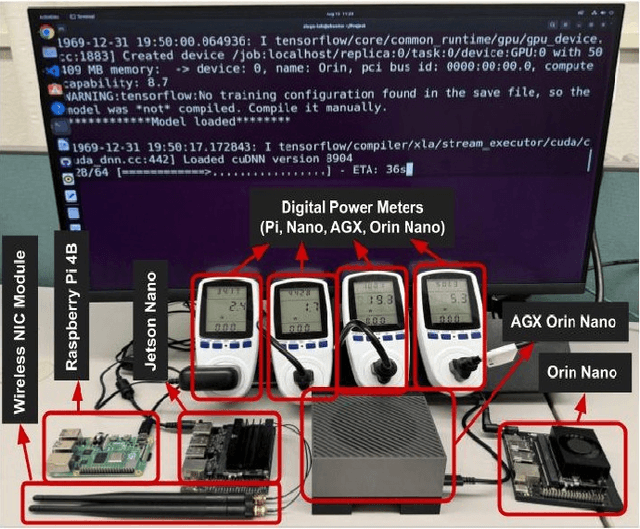
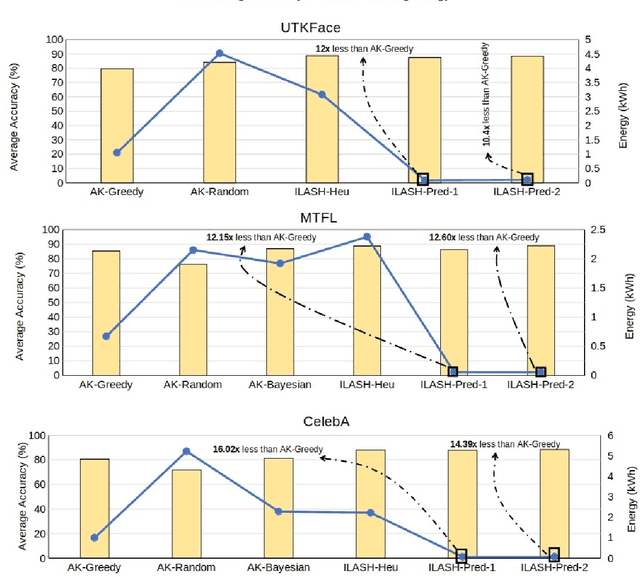
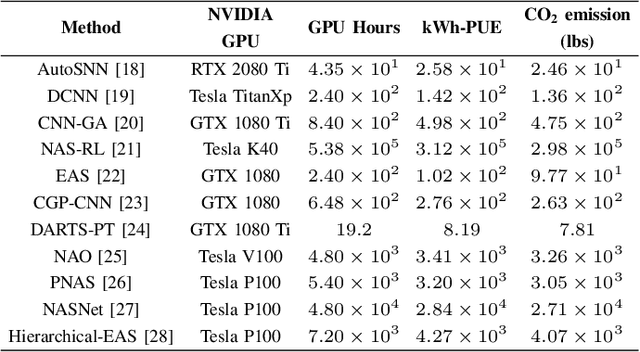
Abstract:Artificial intelligence (AI) is widely used in various fields including healthcare, autonomous vehicles, robotics, traffic monitoring, and agriculture. Many modern AI applications in these fields are multi-tasking in nature (i.e. perform multiple analysis on same data) and are deployed on resource-constrained edge devices requiring the AI models to be efficient across different metrics such as power, frame rate, and size. For these specific use-cases, in this work, we propose a new paradigm of neural network architecture (ILASH) that leverages a layer sharing concept for minimizing power utilization, increasing frame rate, and reducing model size. Additionally, we propose a novel neural network architecture search framework (ILASH-NAS) for efficient construction of these neural network models for a given set of tasks and device constraints. The proposed NAS framework utilizes a data-driven intelligent approach to make the search efficient in terms of energy, time, and CO2 emission. We perform extensive evaluations of the proposed layer shared architecture paradigm (ILASH) and the ILASH-NAS framework using four open-source datasets (UTKFace, MTFL, CelebA, and Taskonomy). We compare ILASH-NAS with AutoKeras and observe significant improvement in terms of both the generated model performance and neural search efficiency with up to 16x less energy utilization, CO2 emission, and training/search time.
LeMo-NADe: Multi-Parameter Neural Architecture Discovery with LLMs
Feb 28, 2024



Abstract:Building efficient neural network architectures can be a time-consuming task requiring extensive expert knowledge. This task becomes particularly challenging for edge devices because one has to consider parameters such as power consumption during inferencing, model size, inferencing speed, and CO2 emissions. In this article, we introduce a novel framework designed to automatically discover new neural network architectures based on user-defined parameters, an expert system, and an LLM trained on a large amount of open-domain knowledge. The introduced framework (LeMo-NADe) is tailored to be used by non-AI experts, does not require a predetermined neural architecture search space, and considers a large set of edge device-specific parameters. We implement and validate this proposed neural architecture discovery framework using CIFAR-10, CIFAR-100, and ImageNet16-120 datasets while using GPT-4 Turbo and Gemini as the LLM component. We observe that the proposed framework can rapidly (within hours) discover intricate neural network models that perform extremely well across a diverse set of application settings defined by the user.
BiofilmScanner: A Computational Intelligence Approach to Obtain Bacterial Cell Morphological Attributes from Biofilm Image
Feb 19, 2023



Abstract:Desulfovibrio alaskensis G20 (DA-G20) is utilized as a model for sulfate-reducing bacteria (SRB) that are associated with corrosion issues caused by microorganisms. SRB-based biofilms are thought to be responsible for the billion-dollar-per-year bio-corrosion of metal infrastructure. Understanding the extraction of the bacterial cells' shape and size properties in the SRB-biofilm at different growth stages will assist with the design of anti-corrosion techniques. However, numerous issues affect current approaches, including time-consuming geometric property extraction, low efficiency, and high error rates. This paper proposes BiofilScanner, a Yolact-based deep learning method integrated with invariant moments to address these problems. Our approach efficiently detects and segments bacterial cells in an SRB image while simultaneously invariant moments measure the geometric characteristics of the segmented cells with low errors. The numerical experiments of the proposed method demonstrate that the BiofilmScanner is 2.1x and 6.8x faster than our earlier Mask-RCNN and DLv3+ methods for detecting, segmenting, and measuring the geometric properties of the cell. Furthermore, the BiofilmScanner achieved an F1-score of 85.28% while Mask-RCNN and DLv3+ obtained F1-scores of 77.67% and 75.18%, respectively.
Hybrid Ant Swarm-Based Data Clustering
Jul 11, 2021



Abstract:Biologically inspired computing techniques are very effective and useful in many areas of research including data clustering. Ant clustering algorithm is a nature-inspired clustering technique which is extensively studied for over two decades. In this study, we extend the ant clustering algorithm (ACA) to a hybrid ant clustering algorithm (hACA). Specifically, we include a genetic algorithm in standard ACA to extend the hybrid algorithm for better performance. We also introduced novel pick up and drop off rules to speed up the clustering performance. We study the performance of the hACA algorithm and compare with standard ACA as a benchmark.
* Conference
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge