Matthias Körschens
Improving Data Efficiency for Plant Cover Prediction with Label Interpolation and Monte-Carlo Cropping
Jul 17, 2023Abstract:The plant community composition is an essential indicator of environmental changes and is, for this reason, usually analyzed in ecological field studies in terms of the so-called plant cover. The manual acquisition of this kind of data is time-consuming, laborious, and prone to human error. Automated camera systems can collect high-resolution images of the surveyed vegetation plots at a high frequency. In combination with subsequent algorithmic analysis, it is possible to objectively extract information on plant community composition quickly and with little human effort. An automated camera system can easily collect the large amounts of image data necessary to train a Deep Learning system for automatic analysis. However, due to the amount of work required to annotate vegetation images with plant cover data, only few labeled samples are available. As automated camera systems can collect many pictures without labels, we introduce an approach to interpolate the sparse labels in the collected vegetation plot time series down to the intermediate dense and unlabeled images to artificially increase our training dataset to seven times its original size. Moreover, we introduce a new method we call Monte-Carlo Cropping. This approach trains on a collection of cropped parts of the training images to deal with high-resolution images efficiently, implicitly augment the training images, and speed up training. We evaluate both approaches on a plant cover dataset containing images of herbaceous plant communities and find that our methods lead to improvements in the species, community, and segmentation metrics investigated.
Domain Adaptation and Active Learning for Fine-Grained Recognition in the Field of Biodiversity
Oct 22, 2021
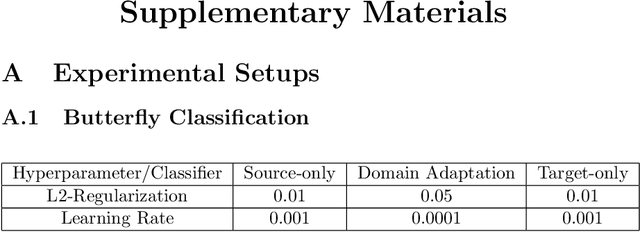

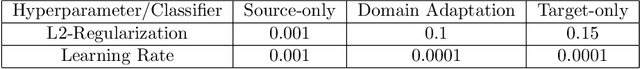
Abstract:Deep-learning methods offer unsurpassed recognition performance in a wide range of domains, including fine-grained recognition tasks. However, in most problem areas there are insufficient annotated training samples. Therefore, the topic of transfer learning respectively domain adaptation is particularly important. In this work, we investigate to what extent unsupervised domain adaptation can be used for fine-grained recognition in a biodiversity context to learn a real-world classifier based on idealized training data, e.g. preserved butterflies and plants. Moreover, we investigate the influence of different normalization layers, such as Group Normalization in combination with Weight Standardization, on the classifier. We discovered that domain adaptation works very well for fine-grained recognition and that the normalization methods have a great influence on the results. Using domain adaptation and Transferable Normalization, the accuracy of the classifier could be increased by up to 12.35 % compared to the baseline. Furthermore, the domain adaptation system is combined with an active learning component to improve the results. We compare different active learning strategies with each other. Surprisingly, we found that more sophisticated strategies provide better results than the random selection baseline for only one of the two datasets. In this case, the distance and diversity strategy performed best. Finally, we present a problem analysis of the datasets.
Automatic Plant Cover Estimation with Convolutional Neural Networks
Jul 02, 2021



Abstract:Monitoring the responses of plants to environmental changes is essential for plant biodiversity research. This, however, is currently still being done manually by botanists in the field. This work is very laborious, and the data obtained is, though following a standardized method to estimate plant coverage, usually subjective and has a coarse temporal resolution. To remedy these caveats, we investigate approaches using convolutional neural networks (CNNs) to automatically extract the relevant data from images, focusing on plant community composition and species coverages of 9 herbaceous plant species. To this end, we investigate several standard CNN architectures and different pretraining methods. We find that we outperform our previous approach at higher image resolutions using a custom CNN with a mean absolute error of 5.16%. In addition to these investigations, we also conduct an error analysis based on the temporal aspect of the plant cover images. This analysis gives insight into where problems for automatic approaches lie, like occlusion and likely misclassifications caused by temporal changes.
Towards Automatic Identification of Elephants in the Wild
Dec 11, 2018



Abstract:Identifying animals from a large group of possible individuals is very important for biodiversity monitoring and especially for collecting data on a small number of particularly interesting individuals, as these have to be identified first before this can be done. Identifying them can be a very time-consuming task. This is especially true, if the animals look very similar and have only a small number of distinctive features, like elephants do. In most cases the animals stay at one place only for a short period of time during which the animal needs to be identified for knowing whether it is important to collect new data on it. For this reason, a system supporting the researchers in identifying elephants to speed up this process would be of great benefit. In this paper, we present such a system for identifying elephants in the face of a large number of individuals with only few training images per individual. For that purpose, we combine object part localization, off-the-shelf CNN features, and support vector machine classification to provide field researches with proposals of possible individuals given new images of an elephant. The performance of our system is demonstrated on a dataset comprising a total of 2078 images of 276 individual elephants, where we achieve 56% top-1 test accuracy and 80% top-10 accuracy. To deal with occlusion, varying viewpoints, and different poses present in the dataset, we furthermore enable the analysts to provide the system with multiple images of the same elephant to be identified and aggregate confidence values generated by the classifier. With that, our system achieves a top-1 accuracy of 74% and a top-10 accuracy of 88% on the held-out test dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge