Matthew Peroni
Deep Trees for (Un)structured Data: Tractability, Performance, and Interpretability
Oct 28, 2024
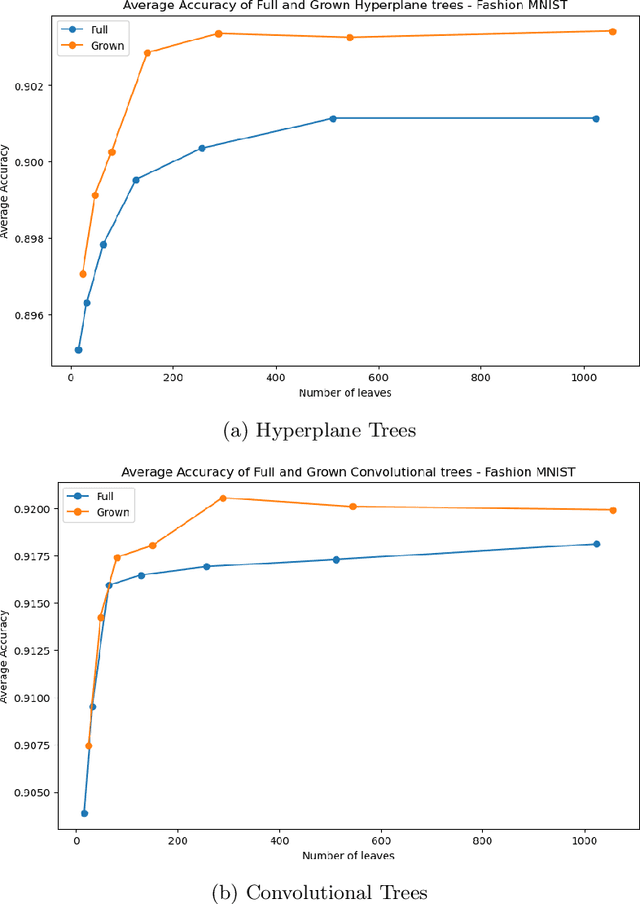
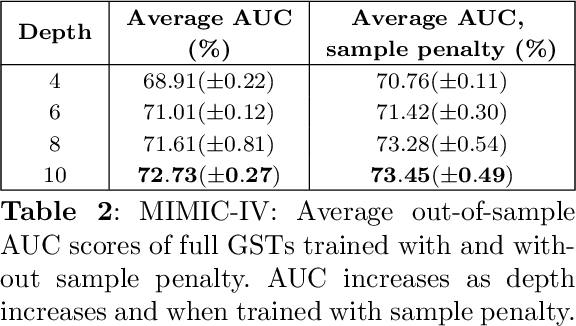
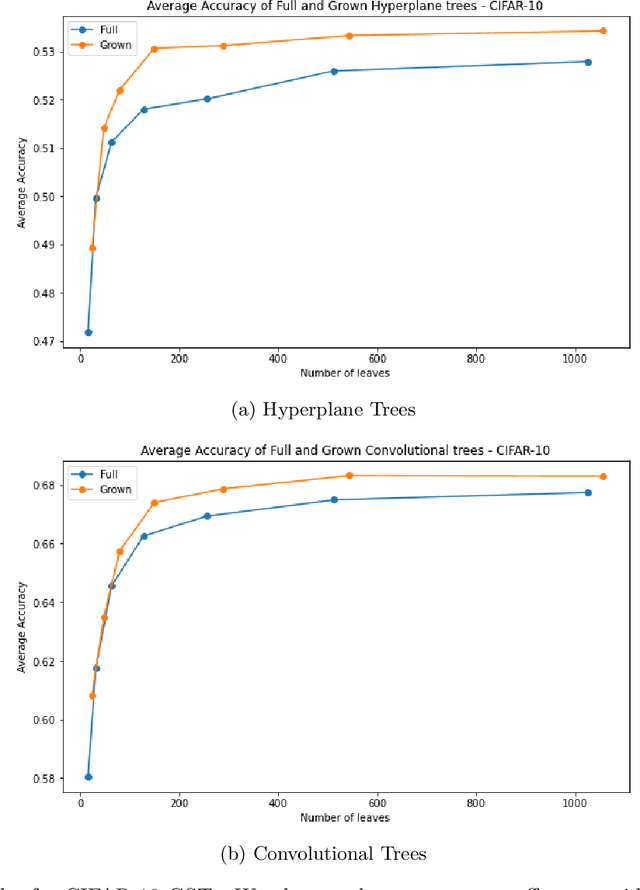
Abstract:Decision Trees have remained a popular machine learning method for tabular datasets, mainly due to their interpretability. However, they lack the expressiveness needed to handle highly nonlinear or unstructured datasets. Motivated by recent advances in tree-based machine learning (ML) techniques and first-order optimization methods, we introduce Generalized Soft Trees (GSTs), which extend soft decision trees (STs) and are capable of processing images directly. We demonstrate their advantages with respect to tractability, performance, and interpretability. We develop a tractable approach to growing GSTs, given by the DeepTree algorithm, which, in addition to new regularization terms, produces high-quality models with far fewer nodes and greater interpretability than traditional soft trees. We test the performance of our GSTs on benchmark tabular and image datasets, including MIMIC-IV, MNIST, Fashion MNIST, CIFAR-10 and Celeb-A. We show that our approach outperforms other popular tree methods (CART, Random Forests, XGBoost) in almost all of the datasets, with Convolutional Trees having a significant edge in the hardest CIFAR-10 and Fashion MNIST datasets. Finally, we explore the interpretability of our GSTs and find that even the most complex GSTs are considerably more interpretable than deep neural networks. Overall, our approach of Generalized Soft Trees provides a tractable method that is high-performing on (un)structured datasets and preserves interpretability more than traditional deep learning methods.
Policy Trees for Prediction: Interpretable and Adaptive Model Selection for Machine Learning
May 30, 2024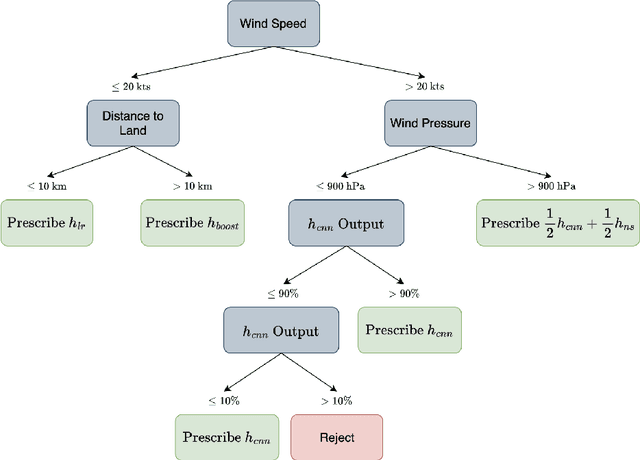
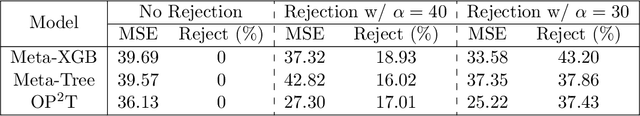
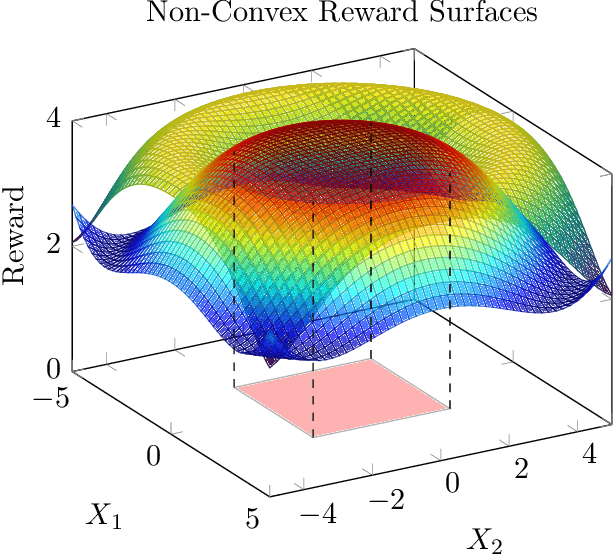
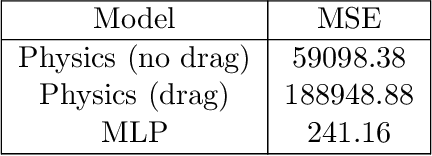
Abstract:As a multitude of capable machine learning (ML) models become widely available in forms such as open-source software and public APIs, central questions remain regarding their use in real-world applications, especially in high-stakes decision-making. Is there always one best model that should be used? When are the models likely to be error-prone? Should a black-box or interpretable model be used? In this work, we develop a prescriptive methodology to address these key questions, introducing a tree-based approach, Optimal Predictive-Policy Trees (OP2T), that yields interpretable policies for adaptively selecting a predictive model or ensemble, along with a parameterized option to reject making a prediction. We base our methods on learning globally optimized prescriptive trees. Our approach enables interpretable and adaptive model selection and rejection while only assuming access to model outputs. By learning policies over different feature spaces, including the model outputs, our approach works with both structured and unstructured datasets. We evaluate our approach on real-world datasets, including regression and classification tasks with both structured and unstructured data. We demonstrate that our approach provides both strong performance against baseline methods while yielding insights that help answer critical questions about which models to use, and when.
Extending the Neural Additive Model for Survival Analysis with EHR Data
Nov 17, 2022Abstract:With increasing interest in applying machine learning to develop healthcare solutions, there is a desire to create interpretable deep learning models for survival analysis. In this paper, we extend the Neural Additive Model (NAM) by incorporating pairwise feature interaction networks and equip these models with loss functions that fit both proportional and non-proportional extensions of the Cox model. We show that within this extended framework, we can construct non-proportional hazard models, which we call TimeNAM, that significantly improve performance over the standard NAM model architecture on benchmark survival datasets. We apply these model architectures to data from the Electronic Health Record (EHR) database of Seoul National University Hospital Gangnam Center (SNUHGC) to build an interpretable neural network survival model for gastric cancer prediction. We demonstrate that on both benchmark survival analysis datasets, as well as on our gastric cancer dataset, our model architectures yield performance that matches, or surpasses, the current state-of-the-art black-box methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge