Masum Shah Junayed
Graph Laplacian Transformer with Progressive Sampling for Prostate Cancer Grading
Dec 11, 2025Abstract:Prostate cancer grading from whole-slide images (WSIs) remains a challenging task due to the large-scale nature of WSIs, the presence of heterogeneous tissue structures, and difficulty of selecting diagnostically relevant regions. Existing approaches often rely on random or static patch selection, leading to the inclusion of redundant or non-informative regions that degrade performance. To address this, we propose a Graph Laplacian Attention-Based Transformer (GLAT) integrated with an Iterative Refinement Module (IRM) to enhance both feature learning and spatial consistency. The IRM iteratively refines patch selection by leveraging a pretrained ResNet50 for local feature extraction and a foundation model in no-gradient mode for importance scoring, ensuring only the most relevant tissue regions are preserved. The GLAT models tissue-level connectivity by constructing a graph where patches serve as nodes, ensuring spatial consistency through graph Laplacian constraints and refining feature representations via a learnable filtering mechanism that enhances discriminative histological structures. Additionally, a convex aggregation mechanism dynamically adjusts patch importance to generate a robust WSI-level representation. Extensive experiments on five public and one private dataset demonstrate that our model outperforms state-of-the-art methods, achieving higher performance and spatial consistency while maintaining computational efficiency.
In-Depth Analysis of Automated Acne Disease Recognition and Classification
Mar 04, 2025Abstract:Facial acne is a common disease, especially among adolescents, negatively affecting both physically and psychologically. Classifying acne is vital to providing the appropriate treatment. Traditional visual inspection or expert scanning is time-consuming and difficult to differentiate acne types. This paper introduces an automated expert system for acne recognition and classification. The proposed method employs a machine learning-based technique to classify and evaluate six types of acne diseases to facilitate the diagnosis of dermatologists. The pre-processing phase includes contrast improvement, smoothing filter, and RGB to L*a*b color conversion to eliminate noise and improve the classification accuracy. Then, a clustering-based segmentation method, k-means clustering, is applied for segmenting the disease-affected regions that pass through the feature extraction step. Characteristics of these disease-affected regions are extracted based on a combination of gray-level co-occurrence matrix (GLCM) and Statistical features. Finally, five different machine learning classifiers are employed to classify acne diseases. Experimental results show that the Random Forest (RF) achieves the highest accuracy of 98.50%, which is promising compared to the state-of-the-art methods.
Multi-modal Spatial Clustering for Spatial Transcriptomics Utilizing High-resolution Histology Images
Oct 31, 2024Abstract:Understanding the intricate cellular environment within biological tissues is crucial for uncovering insights into complex biological functions. While single-cell RNA sequencing has significantly enhanced our understanding of cellular states, it lacks the spatial context necessary to fully comprehend the cellular environment. Spatial transcriptomics (ST) addresses this limitation by enabling transcriptome-wide gene expression profiling while preserving spatial context. One of the principal challenges in ST data analysis is spatial clustering, which reveals spatial domains based on the spots within a tissue. Modern ST sequencing procedures typically include a high-resolution histology image, which has been shown in previous studies to be closely connected to gene expression profiles. However, current spatial clustering methods often fail to fully integrate high-resolution histology image features with gene expression data, limiting their ability to capture critical spatial and cellular interactions. In this study, we propose the spatial transcriptomics multi-modal clustering (stMMC) model, a novel contrastive learning-based deep learning approach that integrates gene expression data with histology image features through a multi-modal parallel graph autoencoder. We tested stMMC against four state-of-the-art baseline models: Leiden, GraphST, SpaGCN, and stLearn on two public ST datasets with 13 sample slices in total. The experiments demonstrated that stMMC outperforms all the baseline models in terms of ARI and NMI. An ablation study further validated the contributions of contrastive learning and the incorporation of histology image features.
An Efficient End-to-End Deep Neural Network for Interstitial Lung Disease Recognition and Classification
Apr 21, 2022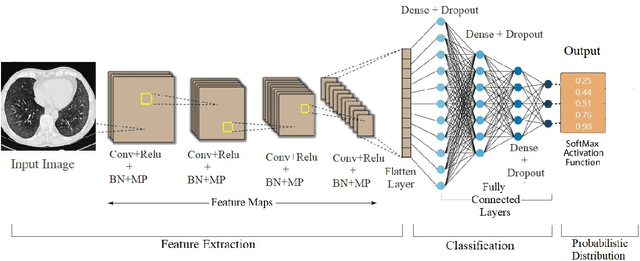
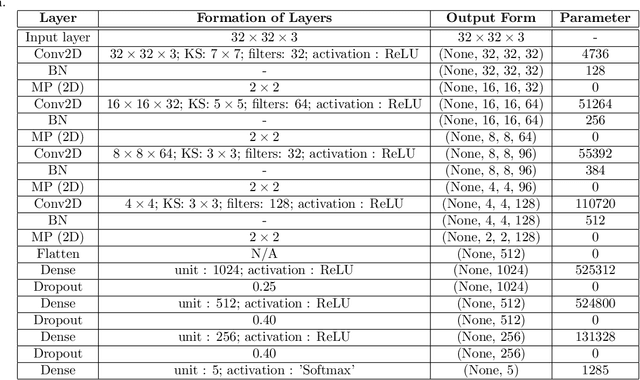
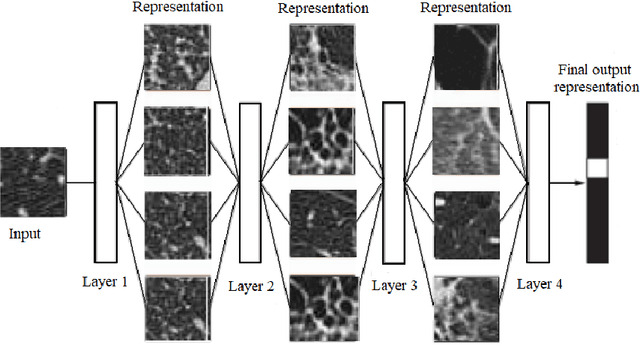
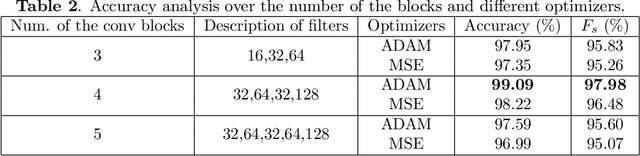
Abstract:The automated Interstitial Lung Diseases (ILDs) classification technique is essential for assisting clinicians during the diagnosis process. Detecting and classifying ILDs patterns is a challenging problem. This paper introduces an end-to-end deep convolution neural network (CNN) for classifying ILDs patterns. The proposed model comprises four convolutional layers with different kernel sizes and Rectified Linear Unit (ReLU) activation function, followed by batch normalization and max-pooling with a size equal to the final feature map size well as four dense layers. We used the ADAM optimizer to minimize categorical cross-entropy. A dataset consisting of 21328 image patches of 128 CT scans with five classes is taken to train and assess the proposed model. A comparison study showed that the presented model outperformed pre-trained CNNs and five-fold cross-validation on the same dataset. For ILDs pattern classification, the proposed approach achieved the accuracy scores of 99.09% and the average F score of 97.9%, outperforming three pre-trained CNNs. These outcomes show that the proposed model is relatively state-of-the-art in precision, recall, f score, and accuracy.
HiMODE: A Hybrid Monocular Omnidirectional Depth Estimation Model
Apr 11, 2022
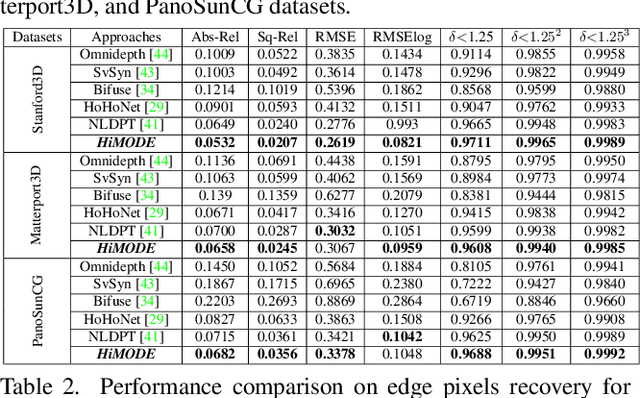
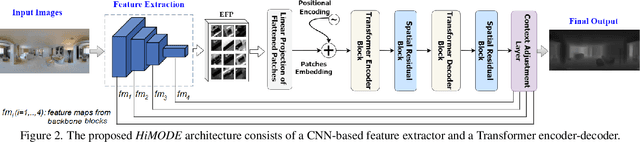
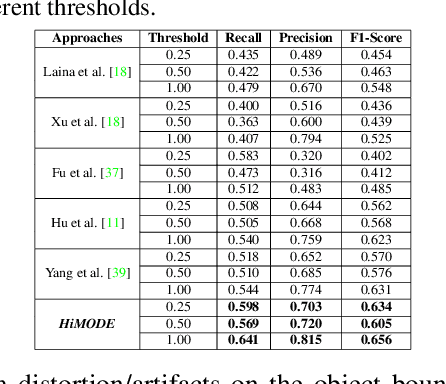
Abstract:Monocular omnidirectional depth estimation is receiving considerable research attention due to its broad applications for sensing 360{\deg} surroundings. Existing approaches in this field suffer from limitations in recovering small object details and data lost during the ground-truth depth map acquisition. In this paper, a novel monocular omnidirectional depth estimation model, namely HiMODE is proposed based on a hybrid CNN+Transformer (encoder-decoder) architecture whose modules are efficiently designed to mitigate distortion and computational cost, without performance degradation. Firstly, we design a feature pyramid network based on the HNet block to extract high-resolution features near the edges. The performance is further improved, benefiting from a self and cross attention layer and spatial/temporal patches in the Transformer encoder and decoder, respectively. Besides, a spatial residual block is employed to reduce the number of parameters. By jointly passing the deep features extracted from an input image at each backbone block, along with the raw depth maps predicted by the transformer encoder-decoder, through a context adjustment layer, our model can produce resulting depth maps with better visual quality than the ground-truth. Comprehensive ablation studies demonstrate the significance of each individual module. Extensive experiments conducted on three datasets; Stanford3D, Matterport3D, and SunCG, demonstrate that HiMODE can achieve state-of-the-art performance for 360{\deg} monocular depth estimation.
Incept-N: A Convolutional Neural Network based Classification Approach for Predicting Nationality from Facial Features
May 18, 2018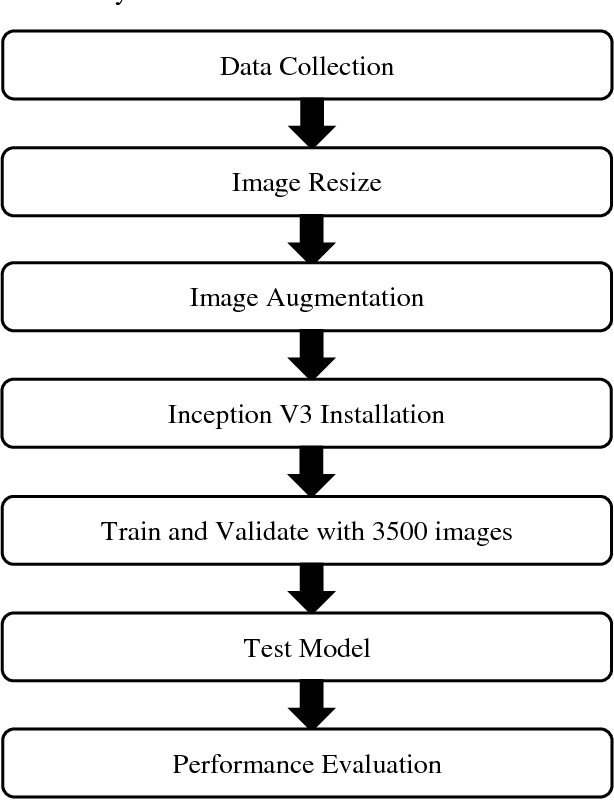

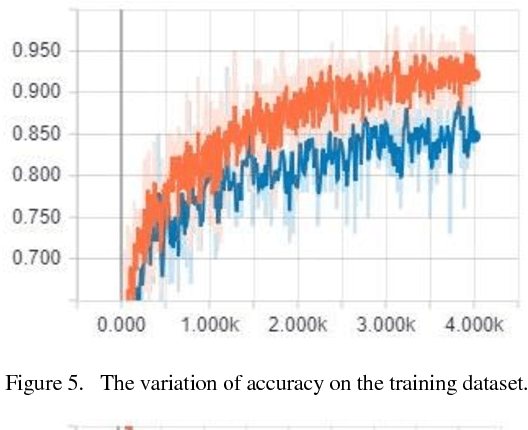
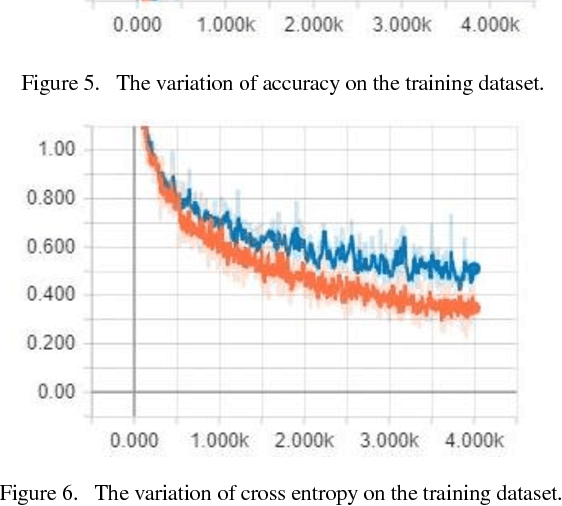
Abstract:The nationality of a human being is a well-known identifying characteristic used for every major authentication purpose in every country. Albeit advances in the application of Artificial Intelligence and Computer Vision in different aspects, its contribution to this specific security procedure is yet to be cultivated. With a goal to successfully applying computer vision techniques to predict the nationality of a person based on his facial features, we have proposed this novel method and have achieved an average of 93.6% accuracy with very low misclassification rate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge