Martin Simonovsky
Deep Learning on Attributed Graphs: A Journey from Graphs to Their Embeddings and Back
Jan 24, 2019



Abstract:A graph is a powerful concept for representation of relations between pairs of entities. Data with underlying graph structure can be found across many disciplines and there is a natural desire for understanding such data better. Deep learning (DL) has achieved significant breakthroughs in a variety of machine learning tasks in recent years, especially where data is structured on a grid, such as in text, speech, or image understanding. However, surprisingly little has been done to explore the applicability of DL on arbitrary graph-structured data directly. The goal of this thesis is to investigate architectures for DL on graphs and study how to transfer, adapt or generalize concepts that work well on sequential and image data to this domain. We concentrate on two important primitives: embedding graphs or their nodes into a continuous vector space representation (encoding) and, conversely, generating graphs from such vectors back (decoding). To that end, we make the following contributions. First, we introduce Edge-Conditioned Convolutions (ECC), a convolution-like operation on graphs performed in the spatial domain where filters are dynamically generated based on edge attributes. The method is used to encode graphs with arbitrary and varying structure. Second, we propose SuperPoint Graph, an intermediate point cloud representation with rich edge attributes encoding the contextual relationship between object parts. Based on this representation, ECC is employed to segment large-scale point clouds without major sacrifice in fine details. Third, we present GraphVAE, a graph generator allowing us to decode graphs with variable but upper-bounded number of nodes making use of approximate graph matching for aligning the predictions of an autoencoder with its inputs. The method is applied to the task of molecule generation.
Large-scale Point Cloud Semantic Segmentation with Superpoint Graphs
Mar 28, 2018
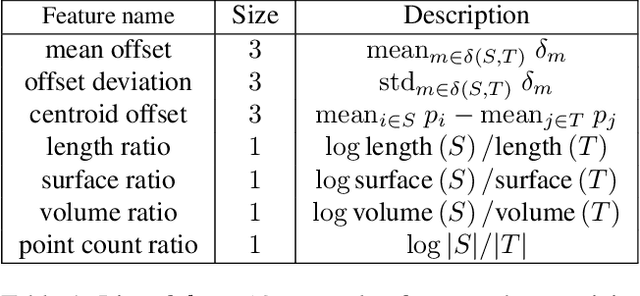
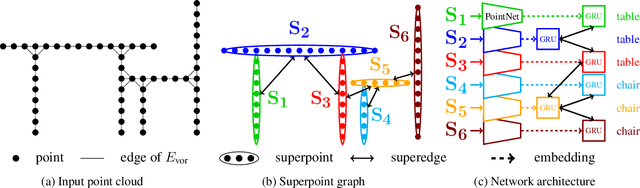
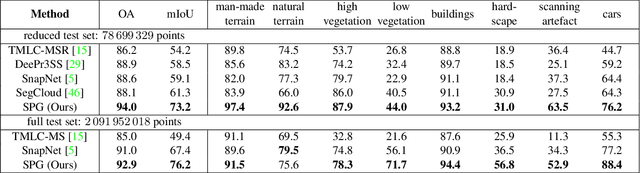
Abstract:We propose a novel deep learning-based framework to tackle the challenge of semantic segmentation of large-scale point clouds of millions of points. We argue that the organization of 3D point clouds can be efficiently captured by a structure called superpoint graph (SPG), derived from a partition of the scanned scene into geometrically homogeneous elements. SPGs offer a compact yet rich representation of contextual relationships between object parts, which is then exploited by a graph convolutional network. Our framework sets a new state of the art for segmenting outdoor LiDAR scans (+11.9 and +8.8 mIoU points for both Semantic3D test sets), as well as indoor scans (+12.4 mIoU points for the S3DIS dataset).
GraphVAE: Towards Generation of Small Graphs Using Variational Autoencoders
Feb 09, 2018


Abstract:Deep learning on graphs has become a popular research topic with many applications. However, past work has concentrated on learning graph embedding tasks, which is in contrast with advances in generative models for images and text. Is it possible to transfer this progress to the domain of graphs? We propose to sidestep hurdles associated with linearization of such discrete structures by having a decoder output a probabilistic fully-connected graph of a predefined maximum size directly at once. Our method is formulated as a variational autoencoder. We evaluate on the challenging task of molecule generation.
Dynamic Edge-Conditioned Filters in Convolutional Neural Networks on Graphs
Aug 08, 2017



Abstract:A number of problems can be formulated as prediction on graph-structured data. In this work, we generalize the convolution operator from regular grids to arbitrary graphs while avoiding the spectral domain, which allows us to handle graphs of varying size and connectivity. To move beyond a simple diffusion, filter weights are conditioned on the specific edge labels in the neighborhood of a vertex. Together with the proper choice of graph coarsening, we explore constructing deep neural networks for graph classification. In particular, we demonstrate the generality of our formulation in point cloud classification, where we set the new state of the art, and on a graph classification dataset, where we outperform other deep learning approaches. The source code is available at https://github.com/mys007/ecc
A Deep Metric for Multimodal Registration
Sep 17, 2016



Abstract:Multimodal registration is a challenging problem in medical imaging due the high variability of tissue appearance under different imaging modalities. The crucial component here is the choice of the right similarity measure. We make a step towards a general learning-based solution that can be adapted to specific situations and present a metric based on a convolutional neural network. Our network can be trained from scratch even from a few aligned image pairs. The metric is validated on intersubject deformable registration on a dataset different from the one used for training, demonstrating good generalization. In this task, we outperform mutual information by a significant margin.
OnionNet: Sharing Features in Cascaded Deep Classifiers
Aug 09, 2016



Abstract:The focus of our work is speeding up evaluation of deep neural networks in retrieval scenarios, where conventional architectures may spend too much time on negative examples. We propose to replace a monolithic network with our novel cascade of feature-sharing deep classifiers, called OnionNet, where subsequent stages may add both new layers as well as new feature channels to the previous ones. Importantly, intermediate feature maps are shared among classifiers, preventing them from the necessity of being recomputed. To accomplish this, the model is trained end-to-end in a principled way under a joint loss. We validate our approach in theory and on a synthetic benchmark. As a result demonstrated in three applications (patch matching, object detection, and image retrieval), our cascade can operate significantly faster than both monolithic networks and traditional cascades without sharing at the cost of marginal decrease in precision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge