Markus Kreuzthaler
Zero- and Few-shot Named Entity Recognition and Text Expansion in Medication Prescriptions using ChatGPT
Sep 26, 2024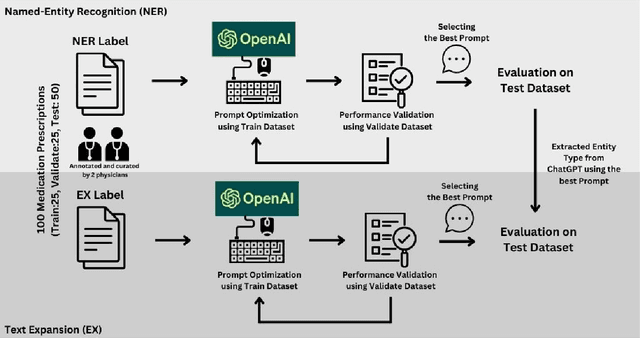
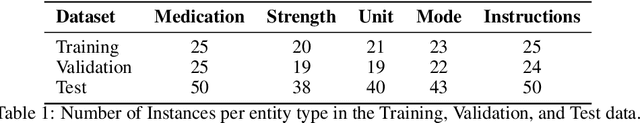
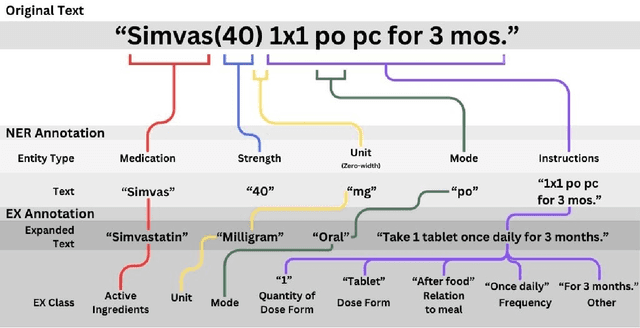
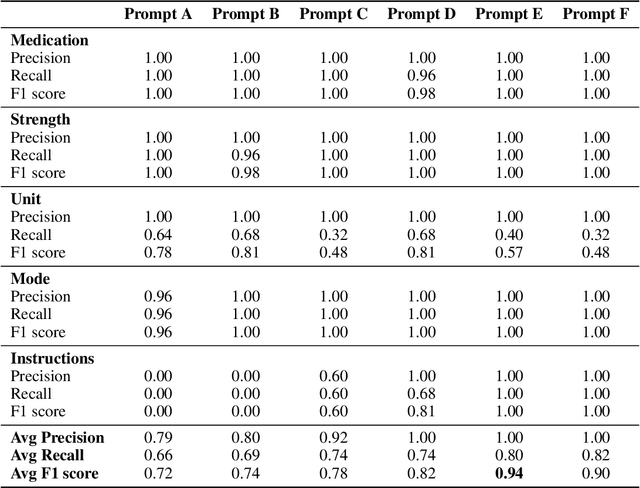
Abstract:Introduction: Medication prescriptions are often in free text and include a mix of two languages, local brand names, and a wide range of idiosyncratic formats and abbreviations. Large language models (LLMs) have shown promising ability to generate text in response to input prompts. We use ChatGPT 3.5 to automatically structure and expand medication statements in discharge summaries and thus make them easier to interpret for people and machines. Methods: Named-entity Recognition (NER) and Text Expansion (EX) are used in a zero- and few-shot setting with different prompt strategies. 100 medication statements were manually annotated and curated. NER performance was measured by using strict and partial matching. For the task EX, two experts interpreted the results by assessing semantic equivalence between original and expanded statements. The model performance was measured by precision, recall, and F1 score. Results: For NER, the best-performing prompt reached an average F1 score of 0.94 in the test set. For EX, the few-shot prompt showed superior performance among other prompts, with an average F1 score of 0.87. Conclusion: Our study demonstrates good performance for NER and EX tasks in free-text medication statements using ChatGPT. Compared to a zero-shot baseline, a few-shot approach prevented the system from hallucinating, which would be unacceptable when processing safety-relevant medication data.
Federated unsupervised random forest for privacy-preserving patient stratification
Jan 29, 2024Abstract:In the realm of precision medicine, effective patient stratification and disease subtyping demand innovative methodologies tailored for multi-omics data. Clustering techniques applied to multi-omics data have become instrumental in identifying distinct subgroups of patients, enabling a finer-grained understanding of disease variability. This work establishes a powerful framework for advancing precision medicine through unsupervised random-forest-based clustering and federated computing. We introduce a novel multi-omics clustering approach utilizing unsupervised random-forests. The unsupervised nature of the random forest enables the determination of cluster-specific feature importance, unraveling key molecular contributors to distinct patient groups. Moreover, our methodology is designed for federated execution, a crucial aspect in the medical domain where privacy concerns are paramount. We have validated our approach on machine learning benchmark data sets as well as on cancer data from The Cancer Genome Atlas (TCGA). Our method is competitive with the state-of-the-art in terms of disease subtyping, but at the same time substantially improves the cluster interpretability. Experiments indicate that local clustering performance can be improved through federated computing.
Secondary Use of Clinical Problem List Entries for Neural Network-Based Disease Code Assignment
Dec 27, 2021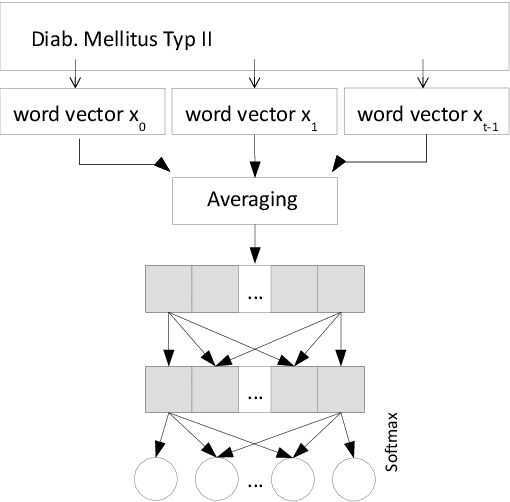
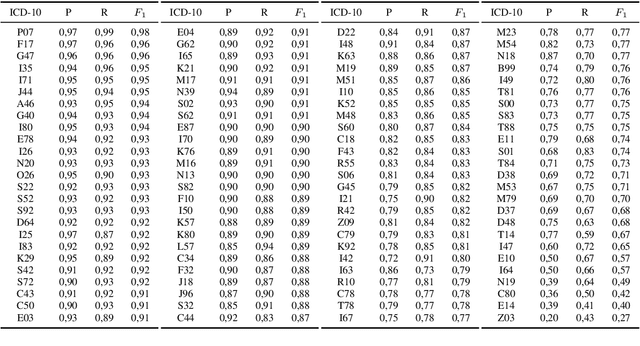
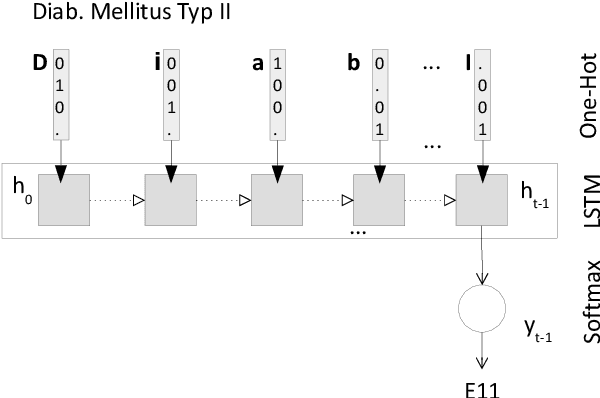
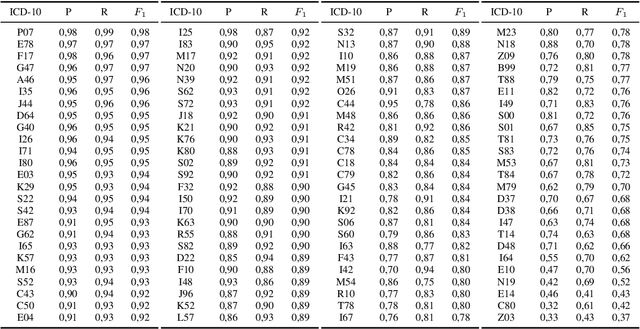
Abstract:Clinical information systems have become large repositories for semi-structured annotated healthcare data, which have reached a critical mass that makes them interesting for supervised data-driven neural network approaches. We explored automated coding of 50 character long clinical problem list entries using the International Classification of Diseases (ICD-10) and evaluated three different types of network architectures on the top 100 ICD-10 three-digit codes. A fastText baseline reached a macro-averaged F1-measure of 0.83, followed by a character-level LSTM with a macro-averaged F1-measure of 0.84. Top performing was a downstreamed RoBERTa model using a custom language model with a macro-averaged F1-measure of 0.88. A neural network activation analysis together with an investigation of the false positives and false negatives unveiled inconsistent manual coding as a main limiting factor.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge