Marcus D. Bloice
Federated unsupervised random forest for privacy-preserving patient stratification
Jan 29, 2024Abstract:In the realm of precision medicine, effective patient stratification and disease subtyping demand innovative methodologies tailored for multi-omics data. Clustering techniques applied to multi-omics data have become instrumental in identifying distinct subgroups of patients, enabling a finer-grained understanding of disease variability. This work establishes a powerful framework for advancing precision medicine through unsupervised random-forest-based clustering and federated computing. We introduce a novel multi-omics clustering approach utilizing unsupervised random-forests. The unsupervised nature of the random forest enables the determination of cluster-specific feature importance, unraveling key molecular contributors to distinct patient groups. Moreover, our methodology is designed for federated execution, a crucial aspect in the medical domain where privacy concerns are paramount. We have validated our approach on machine learning benchmark data sets as well as on cancer data from The Cancer Genome Atlas (TCGA). Our method is competitive with the state-of-the-art in terms of disease subtyping, but at the same time substantially improves the cluster interpretability. Experiments indicate that local clustering performance can be improved through federated computing.
Parea: multi-view ensemble clustering for cancer subtype discovery
Sep 30, 2022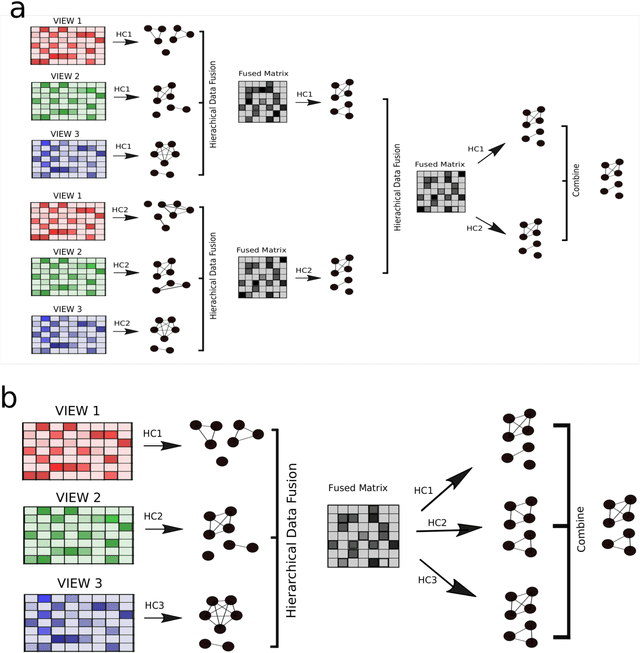
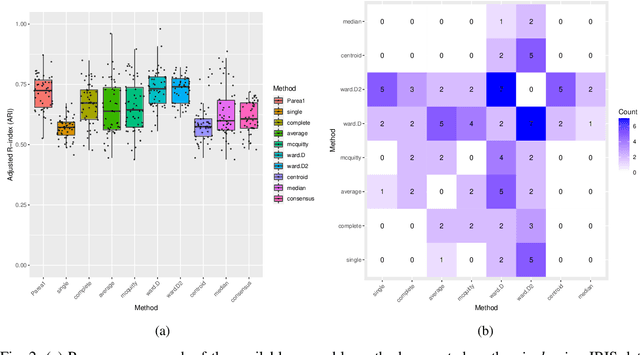
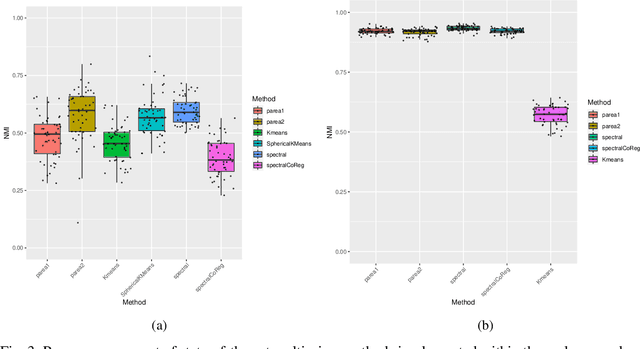
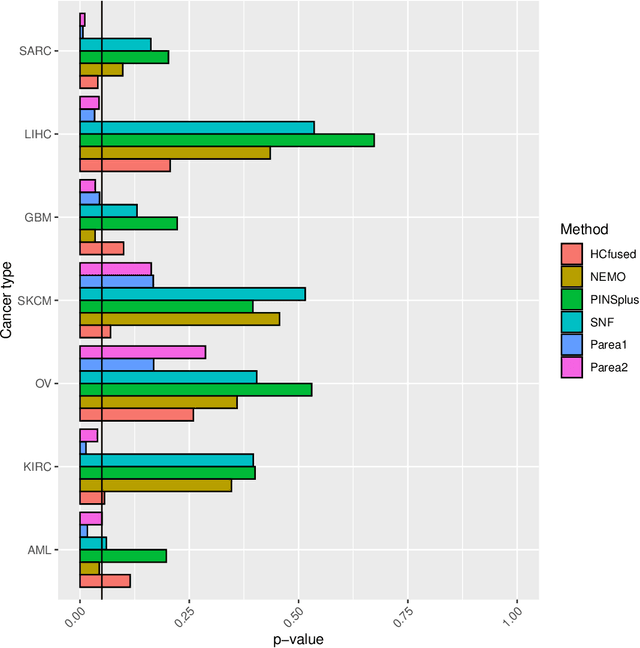
Abstract:Multi-view clustering methods are essential for the stratification of patients into sub-groups of similar molecular characteristics. In recent years, a wide range of methods has been developed for this purpose. However, due to the high diversity of cancer-related data, a single method may not perform sufficiently well in all cases. We present Parea, a multi-view hierarchical ensemble clustering approach for disease subtype discovery. We demonstrate its performance on several machine learning benchmark datasets. We apply and validate our methodology on real-world multi-view cancer patient data. Parea outperforms the current state-of-the-art on six out of seven analysed cancer types. We have integrated the Parea method into our developed Python package Pyrea (https://github.com/mdbloice/Pyrea), which enables the effortless and flexible design of ensemble workflows while incorporating a wide range of fusion and clustering algorithms.
Performing Arithmetic Using a Neural Network Trained on Digit Permutation Pairs
Dec 06, 2019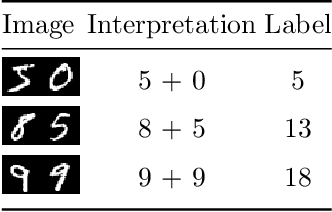
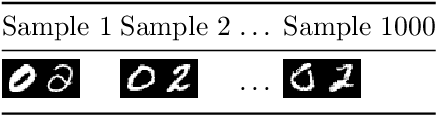
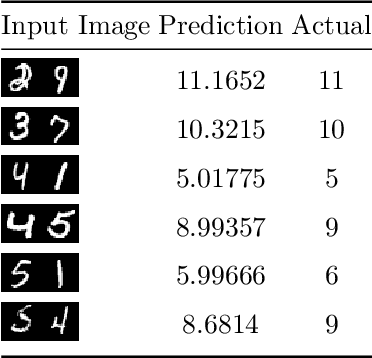
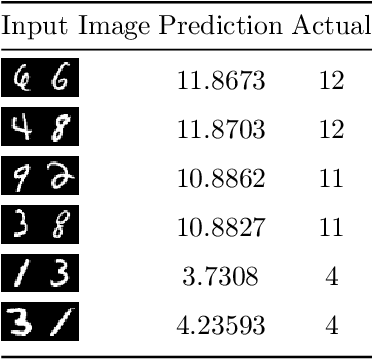
Abstract:In this paper a neural network is trained to perform simple arithmetic using images of concatenated handwritten digit pairs. A convolutional neural network was trained with images consisting of two side-by-side handwritten digits, where the image's label is the summation of the two digits contained in the combined image. Crucially, the network was tested on permutation pairs that were not present during training in an effort to see if the network could learn the task of addition, as opposed to simply mapping images to labels. A dataset was generated for all possible permutation pairs of length 2 for the digits 0-9 using MNIST as a basis for the images, with one thousand samples generated for each permutation pair. For testing the network, samples generated from previously unseen permutation pairs were fed into the trained network, and its predictions measured. Results were encouraging, with the network achieving an accuracy of over 90% on some permutation train/test splits. This suggests that the network learned at first digit recognition, and subsequently the further task of addition based on the two recognised digits. As far as the authors are aware, no previous work has concentrated on learning a mathematical operation in this way.
Patch augmentation: Towards efficient decision boundaries for neural networks
Nov 25, 2019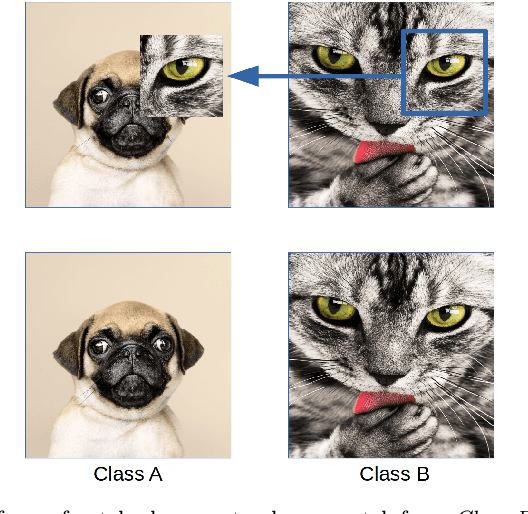
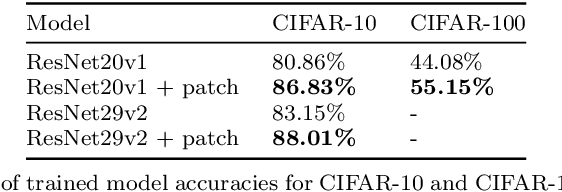
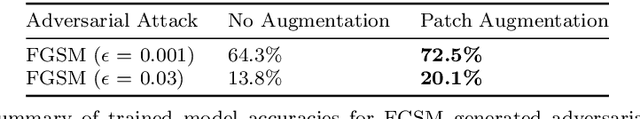
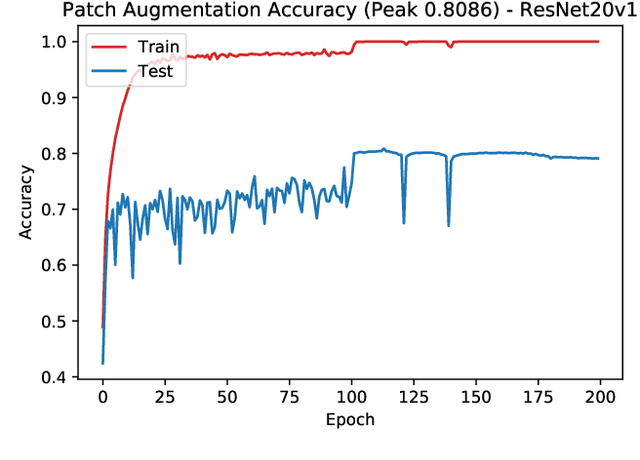
Abstract:In this paper we propose a new augmentation technique, called patch augmentation, that, in our experiments, improves model accuracy and makes networks more robust to adversarial attacks. In brief, this data-independent approach creates new image data based on image/label pairs, where a patch from one of the two images in the pair is superimposed on to the other image, creating a new augmented sample. The new image's label is a linear combination of the image pair's corresponding labels. Initial experiments show a several percentage point increase in accuracy on CIFAR-10, from a baseline of approximately 81% to 89%. CIFAR-100 sees larger improvements still, from a baseline of 52% to 68% accuracy. Networks trained using patch augmentation are also more robust to adversarial attacks, which we demonstrate using the Fast Gradient Sign Method.
Augmentor: An Image Augmentation Library for Machine Learning
Aug 11, 2017

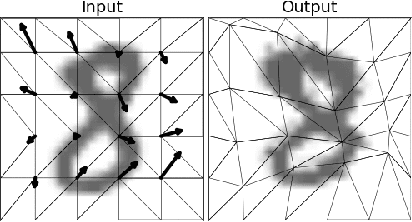
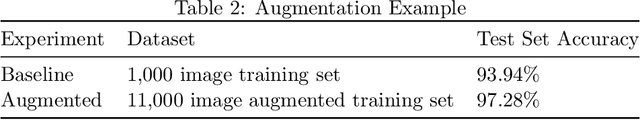
Abstract:The generation of artificial data based on existing observations, known as data augmentation, is a technique used in machine learning to improve model accuracy, generalisation, and to control overfitting. Augmentor is a software package, available in both Python and Julia versions, that provides a high level API for the expansion of image data using a stochastic, pipeline-based approach which effectively allows for images to be sampled from a distribution of augmented images at runtime. Augmentor provides methods for most standard augmentation practices as well as several advanced features such as label-preserving, randomised elastic distortions, and provides many helper functions for typical augmentation tasks used in machine learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge