Mark Basham
PERC: a suite of software tools for the curation of cryoEM data with application to simulation, modelling and machine learning
Mar 17, 2025Abstract:Ease of access to data, tools and models expedites scientific research. In structural biology there are now numerous open repositories of experimental and simulated datasets. Being able to easily access and utilise these is crucial for allowing researchers to make optimal use of their research effort. The tools presented here are useful for collating existing public cryoEM datasets and/or creating new synthetic cryoEM datasets to aid the development of novel data processing and interpretation algorithms. In recent years, structural biology has seen the development of a multitude of machine-learning based algorithms for aiding numerous steps in the processing and reconstruction of experimental datasets and the use of these approaches has become widespread. Developing such techniques in structural biology requires access to large datasets which can be cumbersome to curate and unwieldy to make use of. In this paper we present a suite of Python software packages which we collectively refer to as PERC (profet, EMPIARreader and CAKED). These are designed to reduce the burden which data curation places upon structural biology research. The protein structure fetcher (profet) package allows users to conveniently download and cleave sequences or structures from the Protein Data Bank or Alphafold databases. EMPIARreader allows lazy loading of Electron Microscopy Public Image Archive datasets in a machine-learning compatible structure. The Class Aggregator for Key Electron-microscopy Data (CAKED) package is designed to seamlessly facilitate the training of machine learning models on electron microscopy data, including electron-cryo-microscopy-specific data augmentation and labelling. These packages may be utilised independently or as building blocks in workflows. All are available in open source repositories and designed to be easily extensible to facilitate more advanced workflows if required.
Affinity-VAE for disentanglement, clustering and classification of objects in multidimensional image data
Sep 09, 2022

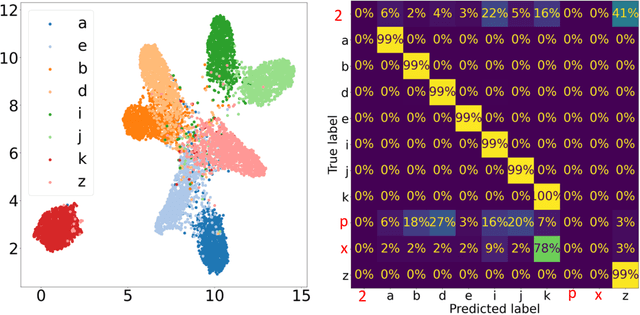

Abstract:In this work we present affinity-VAE: a framework for automatic clustering and classification of objects in multidimensional image data based on their similarity. The method expands on the concept of $\beta$-VAEs with an informed similarity-based loss component driven by an affinity matrix. The affinity-VAE is able to create rotationally-invariant, morphologically homogeneous clusters in the latent representation, with improved cluster separation compared with a standard $\beta$-VAE. We explore the extent of latent disentanglement and continuity of the latent spaces on both 2D and 3D image data, including simulated biological electron cryo-tomography (cryo-ET) volumes as an example of a scientific application.
Savu: A Python-based, MPI Framework for Simultaneous Processing of Multiple, N-dimensional, Large Tomography Datasets
Oct 24, 2016
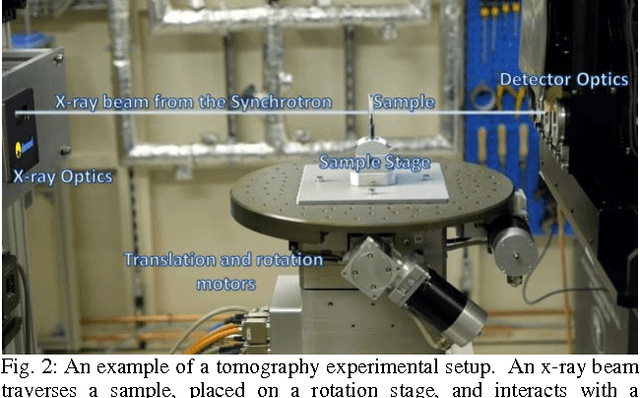
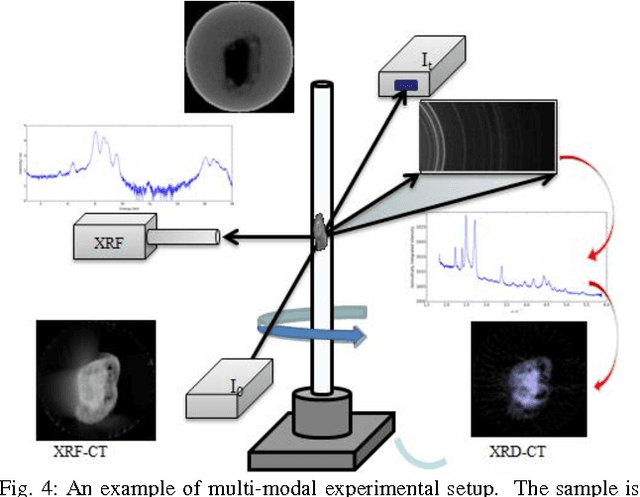

Abstract:Diamond Light Source (DLS), the UK synchrotron facility, attracts scientists from across the world to perform ground-breaking x-ray experiments. With over 3000 scientific users per year, vast amounts of data are collected across the experimental beamlines, with the highest volume of data collected during tomographic imaging experiments. A growing interest in tomography as an imaging technique, has led to an expansion in the range of experiments performed, in addition to a growth in the size of the data per experiment. Savu is a portable, flexible, scientific processing pipeline capable of processing multiple, n-dimensional datasets in serial on a PC, or in parallel across a cluster. Developed at DLS, and successfully deployed across the beamlines, it uses a modular plugin format to enable experiment-specific processing and utilises parallel HDF5 to remove RAM restrictions. The Savu design, described throughout this paper, focuses on easy integration of existing and new functionality, flexibility and ease of use for users and developers alike.
Hierarchical Piecewise-Constant Super-regions
May 19, 2016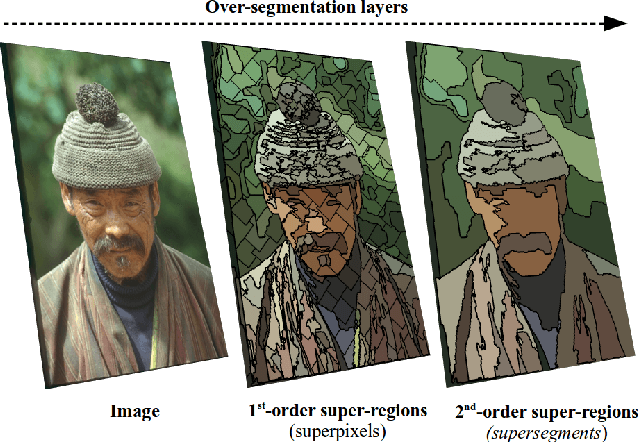
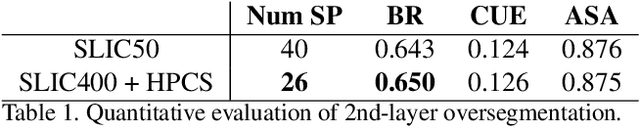

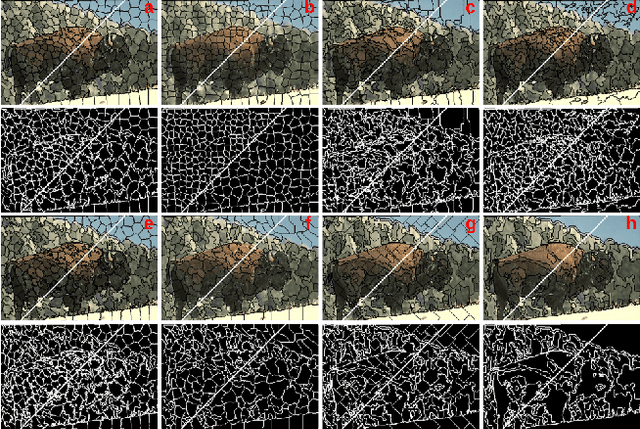
Abstract:Recent applications in computer vision have come to heavily rely on superpixel over-segmentation as a pre-processing step for higher level vision tasks, such as object recognition, image labelling or image segmentation. Here we present a new superpixel algorithm called Hierarchical Piecewise-Constant Super-regions (HPCS), which not only obtains superpixels comparable to the state-of-the-art, but can also be applied hierarchically to form what we call n-th order super-regions. In essence, a Markov Random Field (MRF)-based anisotropic denoising formulation over the quantized feature space is adopted to form piecewise-constant image regions, which are then combined with a graph-based split & merge post-processing step to form superpixels. The graph and quantized feature based formulation of the problem allows us to generalize it hierarchically to preserve boundary adherence with fewer superpixels. Experimental results show that, despite the simplicity of our framework, it is able to provide high quality superpixels, and to hierarchically apply them to form layers of over-segmentation, each with a decreasing number of superpixels, while maintaining the same desired properties (such as adherence to strong image edges). The algorithm is also memory efficient and has a low computational cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge