Marina Pominova
Fader Networks for domain adaptation on fMRI: ABIDE-II study
Oct 14, 2020Abstract:ABIDE is the largest open-source autism spectrum disorder database with both fMRI data and full phenotype description. These data were extensively studied based on functional connectivity analysis as well as with deep learning on raw data, with top models accuracy close to 75\% for separate scanning sites. Yet there is still a problem of models transferability between different scanning sites within ABIDE. In the current paper, we for the first time perform domain adaptation for brain pathology classification problem on raw neuroimaging data. We use 3D convolutional autoencoders to build the domain irrelevant latent space image representation and demonstrate this method to outperform existing approaches on ABIDE data.
Domain Shift in Computer Vision models for MRI data analysis: An Overview
Oct 14, 2020Abstract:Machine learning and computer vision methods are showing good performance in medical imagery analysis. Yetonly a few applications are now in clinical use and one of the reasons for that is poor transferability of themodels to data from different sources or acquisition domains. Development of new methods and algorithms forthe transfer of training and adaptation of the domain in multi-modal medical imaging data is crucial for thedevelopment of accurate models and their use in clinics. In present work, we overview methods used to tackle thedomain shift problem in machine learning and computer vision. The algorithms discussed in this survey includeadvanced data processing, model architecture enhancing and featured training, as well as predicting in domaininvariant latent space. The application of the autoencoding neural networks and their domain-invariant variationsare heavily discussed in a survey. We observe the latest methods applied to the magnetic resonance imaging(MRI) data analysis and conclude on their performance as well as propose directions for further research.
* 8 pages, 1 figure
3D Deformable Convolutions for MRI classification
Nov 05, 2019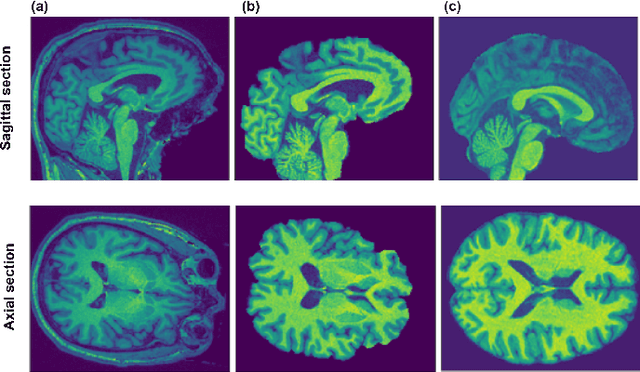
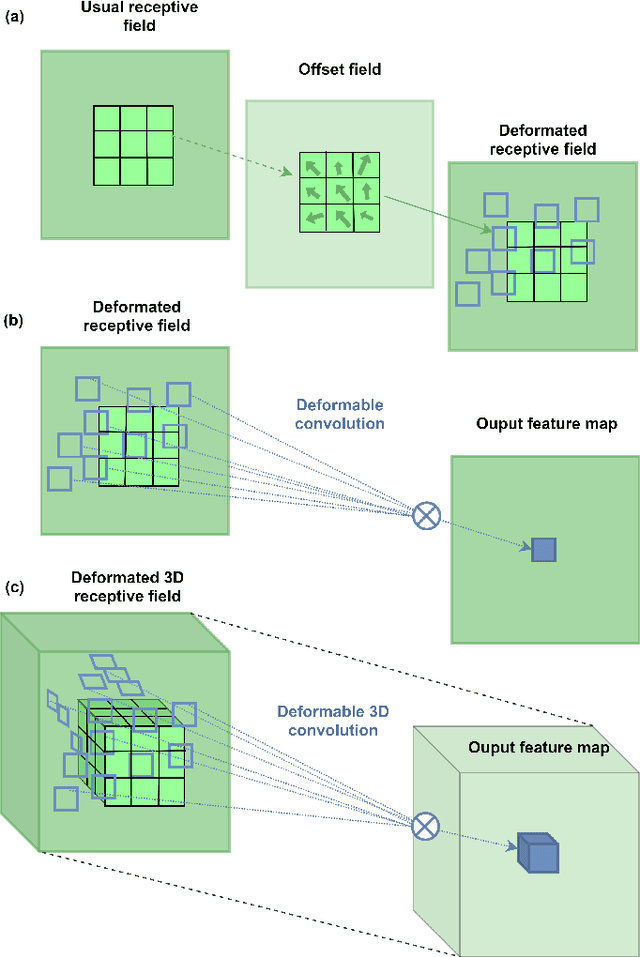
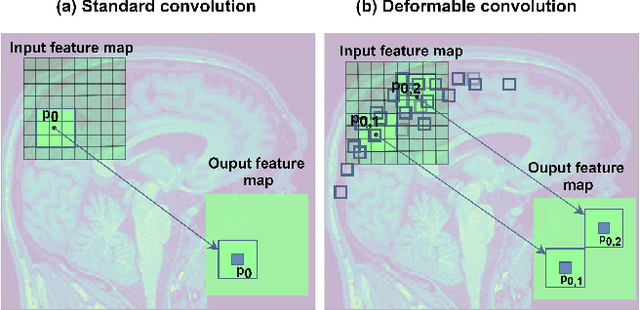
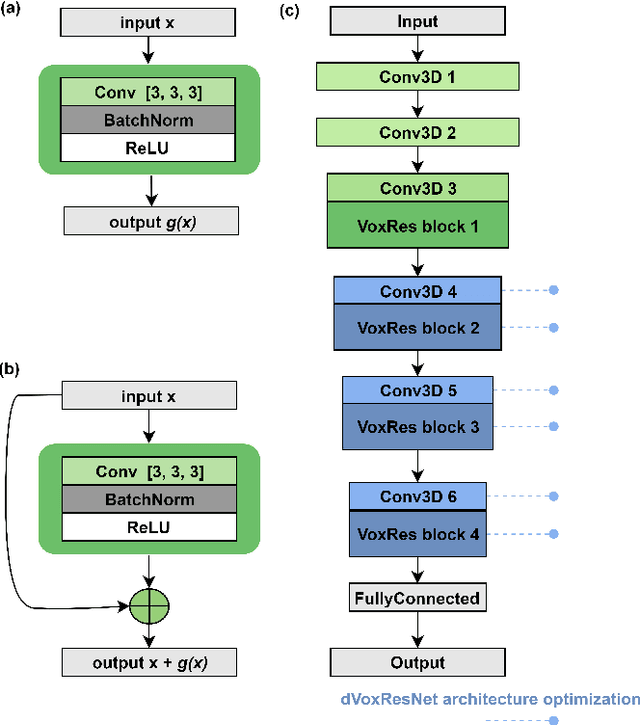
Abstract:Deep learning convolutional neural networks have proved to be a powerful tool for MRI analysis. In current work, we explore the potential of the deformable convolutional deep neural network layers for MRI data classification. We propose new 3D deformable convolutions(d-convolutions), implement them in VoxResNet architecture and apply for structural MRI data classification. We show that 3D d-convolutions outperform standard ones and are effective for unprocessed 3D MR images being robust to particular geometrical properties of the data. Firstly proposed dVoxResNet architecture exhibits high potential for the use in MRI data classification.
Ensemble of 3D CNN regressors with data fusion for fluid intelligence prediction
May 25, 2019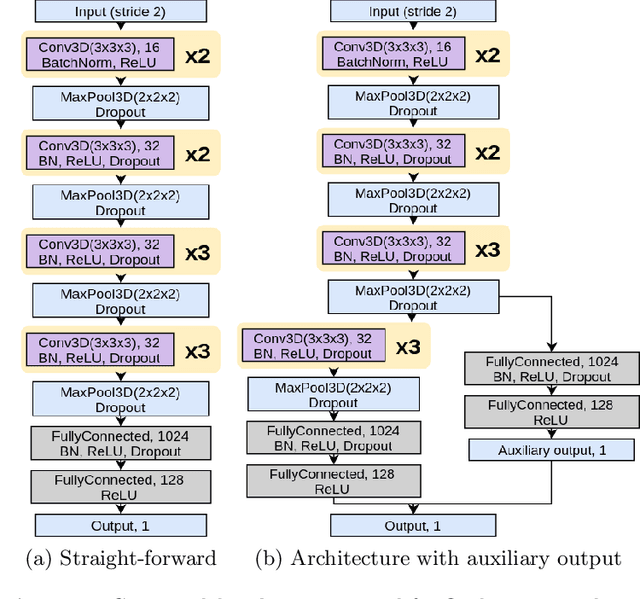


Abstract:In this work, we aim at predicting children's fluid intelligence scores based on structural T1-weighted MR images from the largest long-term study of brain development and child health. The target variable was regressed on a data collection site, socio-demographic variables and brain volume, thus being independent to the potentially informative factors, which are not directly related to the brain functioning. We investigate both feature extraction and deep learning approaches as well as different deep CNN architectures and their ensembles. We propose an advanced architecture of VoxCNNs ensemble, which yield MSE (92.838) on blind test.
* 10 pages, 1 figure, 2 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge