Mariana Neves
Detection of fields of applications in biomedical abstracts with the support of argumentation elements
Apr 09, 2024


Abstract:Focusing on particular facts, instead of the complete text, can potentially improve searching for specific information in the scientific literature. In particular, argumentative elements allow focusing on specific parts of a publication, e.g., the background section or the claims from the authors. We evaluated some tools for the extraction of argumentation elements for a specific task in biomedicine, namely, for detecting the fields of the application in a biomedical publication, e.g, whether it addresses the problem of disease diagnosis or drug development. We performed experiments with the PubMedBERT pre-trained model, which was fine-tuned on a specific corpus for the task. We compared the use of title and abstract to restricting to only some argumentative elements. The top F1 scores ranged from 0.22 to 0.84, depending on the field of application. The best argumentative labels were the ones related the conclusion and background sections of an abstract.
Annotationsaurus: A Searchable Directory of Annotation Tools
Oct 14, 2020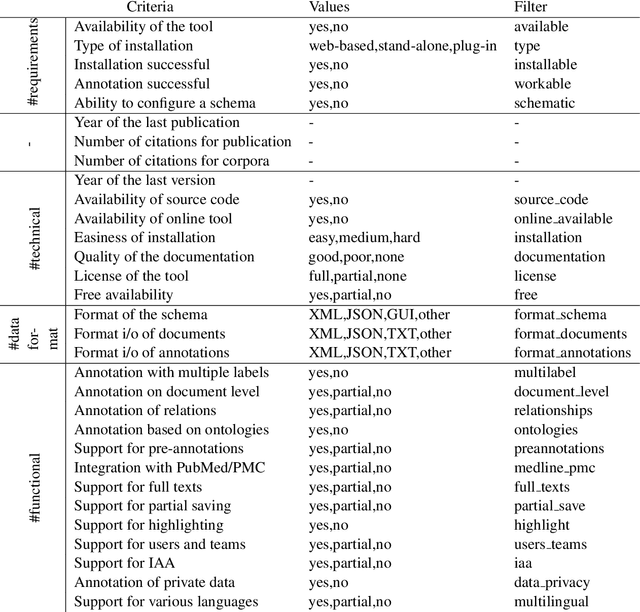
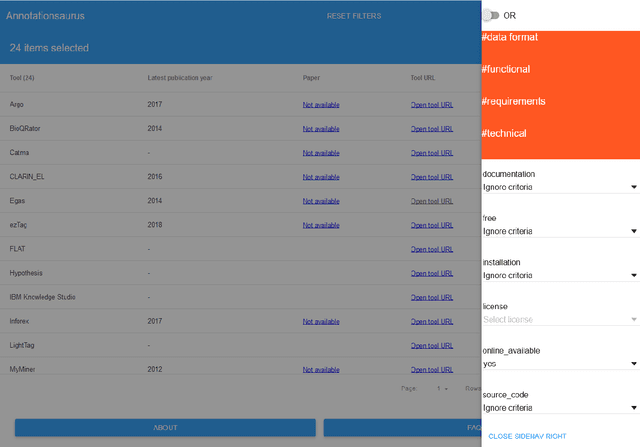
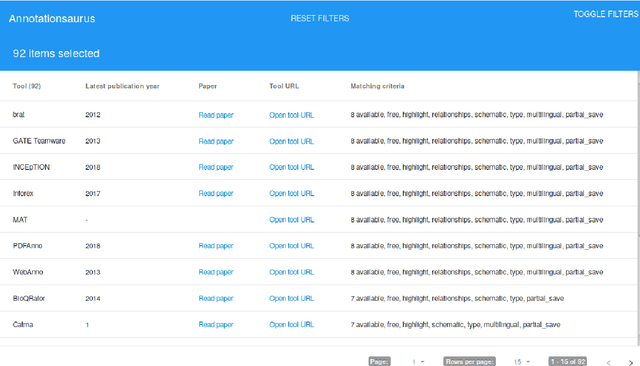
Abstract:Manual annotation of textual documents is a necessary task when constructing benchmark corpora for training and evaluating machine learning algorithms. We created a comprehensive directory of annotation tools that currently includes 93 tools. We analyzed the tools over a set of 31 features and implemented simple scripts and a Web application that filters the tools based on chosen criteria. We present two use cases using the directory and propose ideas for its maintenance. The directory, source codes for scripts, and link to the Web application are available at: https://github.com/mariananeves/annotation-tools
RDF2PT: Generating Brazilian Portuguese Texts from RDF Data
Feb 22, 2018

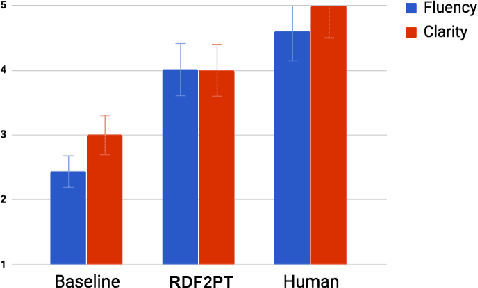
Abstract:The generation of natural language from Resource Description Framework (RDF) data has recently gained significant attention due to the continuous growth of Linked Data. A number of these approaches generate natural language in languages other than English, however, no work has been proposed to generate Brazilian Portuguese texts out of RDF. We address this research gap by presenting RDF2PT, an approach that verbalizes RDF data to Brazilian Portuguese language. We evaluated RDF2PT in an open questionnaire with 44 native speakers divided into experts and non-experts. Our results suggest that RDF2PT is able to generate text which is similar to that generated by humans and can hence be easily understood.
Neural Question Answering at BioASQ 5B
Jun 26, 2017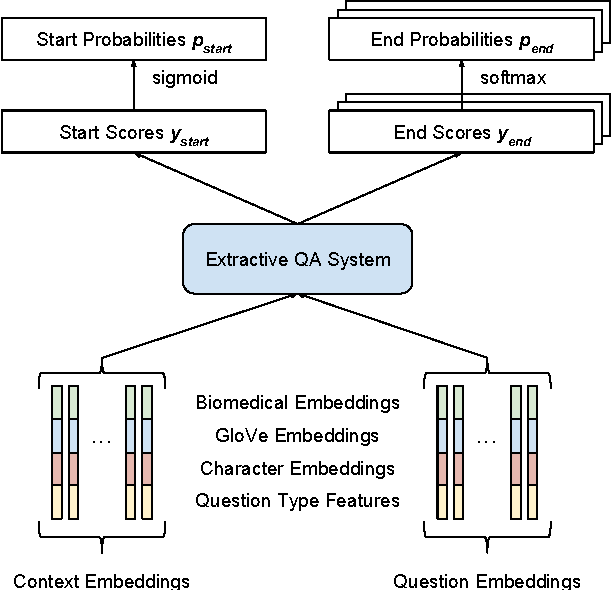
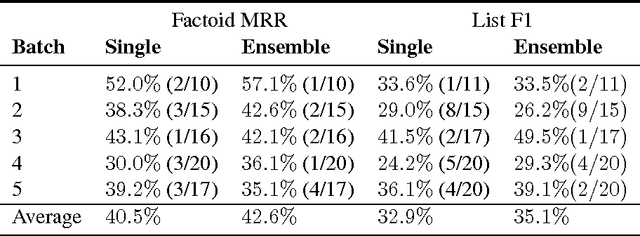
Abstract:This paper describes our submission to the 2017 BioASQ challenge. We participated in Task B, Phase B which is concerned with biomedical question answering (QA). We focus on factoid and list question, using an extractive QA model, that is, we restrict our system to output substrings of the provided text snippets. At the core of our system, we use FastQA, a state-of-the-art neural QA system. We extended it with biomedical word embeddings and changed its answer layer to be able to answer list questions in addition to factoid questions. We pre-trained the model on a large-scale open-domain QA dataset, SQuAD, and then fine-tuned the parameters on the BioASQ training set. With our approach, we achieve state-of-the-art results on factoid questions and competitive results on list questions.
Neural Domain Adaptation for Biomedical Question Answering
Jun 15, 2017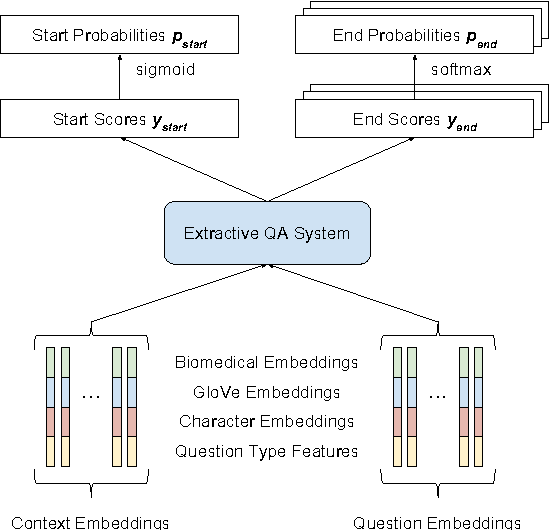
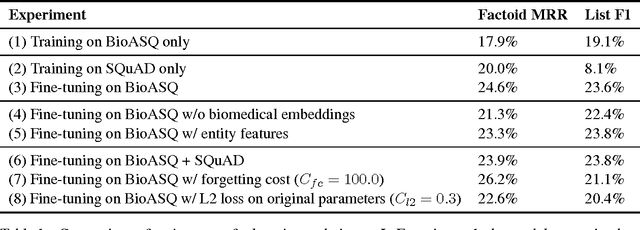
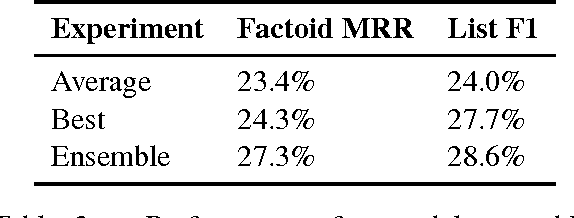
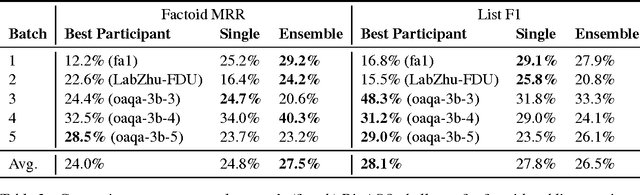
Abstract:Factoid question answering (QA) has recently benefited from the development of deep learning (DL) systems. Neural network models outperform traditional approaches in domains where large datasets exist, such as SQuAD (ca. 100,000 questions) for Wikipedia articles. However, these systems have not yet been applied to QA in more specific domains, such as biomedicine, because datasets are generally too small to train a DL system from scratch. For example, the BioASQ dataset for biomedical QA comprises less then 900 factoid (single answer) and list (multiple answers) QA instances. In this work, we adapt a neural QA system trained on a large open-domain dataset (SQuAD, source) to a biomedical dataset (BioASQ, target) by employing various transfer learning techniques. Our network architecture is based on a state-of-the-art QA system, extended with biomedical word embeddings and a novel mechanism to answer list questions. In contrast to existing biomedical QA systems, our system does not rely on domain-specific ontologies, parsers or entity taggers, which are expensive to create. Despite this fact, our systems achieve state-of-the-art results on factoid questions and competitive results on list questions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge