Mani Valleti
Rapid optimization in high dimensional space by deep kernel learning augmented genetic algorithms
Oct 04, 2024Abstract:Exploration of complex high-dimensional spaces presents significant challenges in fields such as molecular discovery, process optimization, and supply chain management. Genetic Algorithms (GAs), while offering significant power for creating new candidate spaces, often entail high computational demands due to the need for evaluation of each new proposed solution. On the other hand, Deep Kernel Learning (DKL) efficiently navigates the spaces of preselected candidate structures but lacks generative capabilities. This study introduces an approach that amalgamates the generative power of GAs to create new candidates with the efficiency of DKL-based surrogate models to rapidly ascertain the behavior of new candidate spaces. This DKL-GA framework can be further used to build Bayesian Optimization (BO) workflows. We demonstrate the effectiveness of this approach through the optimization of the FerroSIM model, showcasing its broad applicability to diverse challenges, including molecular discovery and battery charging optimization.
Invariant Discovery of Features Across Multiple Length Scales: Applications in Microscopy and Autonomous Materials Characterization
Aug 01, 2024Abstract:Physical imaging is a foundational characterization method in areas from condensed matter physics and chemistry to astronomy and spans length scales from atomic to universe. Images encapsulate crucial data regarding atomic bonding, materials microstructures, and dynamic phenomena such as microstructural evolution and turbulence, among other phenomena. The challenge lies in effectively extracting and interpreting this information. Variational Autoencoders (VAEs) have emerged as powerful tools for identifying underlying factors of variation in image data, providing a systematic approach to distilling meaningful patterns from complex datasets. However, a significant hurdle in their application is the definition and selection of appropriate descriptors reflecting local structure. Here we introduce the scale-invariant VAE approach (SI-VAE) based on the progressive training of the VAE with the descriptors sampled at different length scales. The SI-VAE allows the discovery of the length scale dependent factors of variation in the system. Here, we illustrate this approach using the ferroelectric domain images and generalize it to the movies of the electron-beam induced phenomena in graphene and topography evolution across combinatorial libraries. This approach can further be used to initialize the decision making in automated experiments including structure-property discovery and can be applied across a broad range of imaging methods. This approach is universal and can be applied to any spatially resolved data including both experimental imaging studies and simulations, and can be particularly useful for exploration of phenomena such as turbulence, scale-invariant transformation fronts, etc.
Physics and Chemistry from Parsimonious Representations: Image Analysis via Invariant Variational Autoencoders
Mar 30, 2023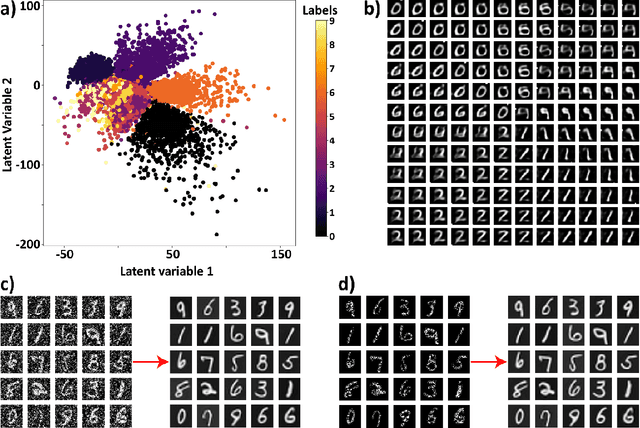
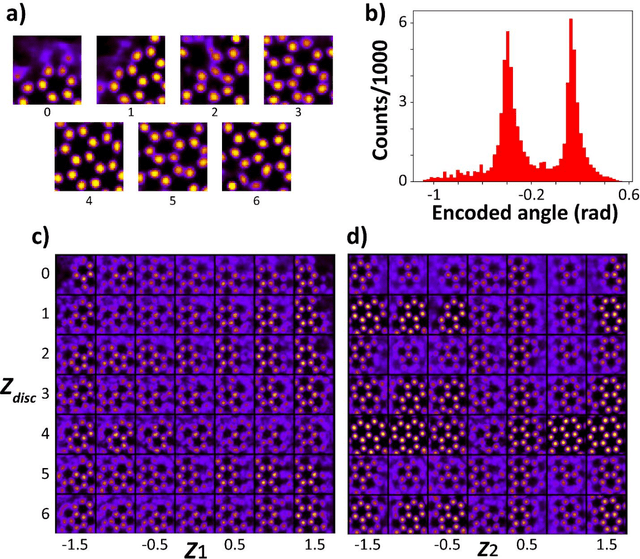
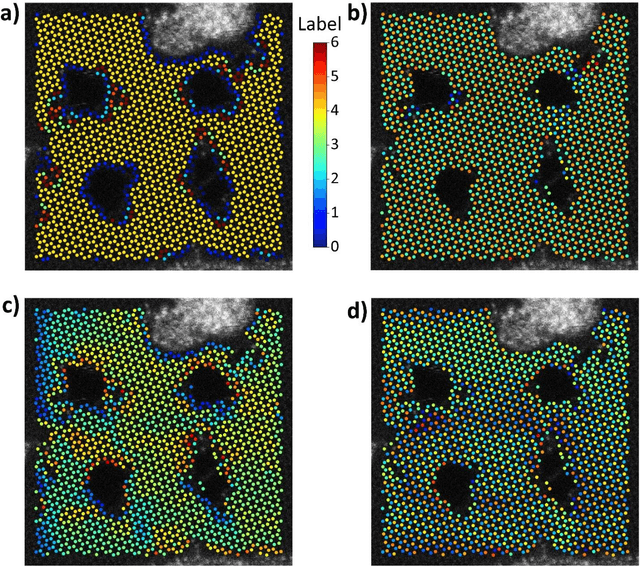
Abstract:Electron, optical, and scanning probe microscopy methods are generating ever increasing volume of image data containing information on atomic and mesoscale structures and functionalities. This necessitates the development of the machine learning methods for discovery of physical and chemical phenomena from the data, such as manifestations of symmetry breaking in electron and scanning tunneling microscopy images, variability of the nanoparticles. Variational autoencoders (VAEs) are emerging as a powerful paradigm for the unsupervised data analysis, allowing to disentangle the factors of variability and discover optimal parsimonious representation. Here, we summarize recent developments in VAEs, covering the basic principles and intuition behind the VAEs. The invariant VAEs are introduced as an approach to accommodate scale and translation invariances present in imaging data and separate known factors of variations from the ones to be discovered. We further describe the opportunities enabled by the control over VAE architecture, including conditional, semi-supervised, and joint VAEs. Several case studies of VAE applications for toy models and experimental data sets in Scanning Transmission Electron Microscopy are discussed, emphasizing the deep connection between VAE and basic physical principles. All the codes used here are available at https://github.com/saimani5/VAE-tutorials and this article can be used as an application guide when applying these to own data sets.
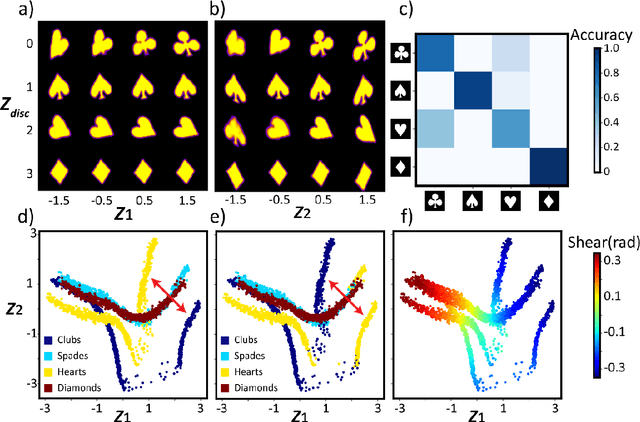
Deep Kernel Methods Learn Better: From Cards to Process Optimization
Mar 25, 2023Abstract:The ability of deep learning methods to perform classification and regression tasks relies heavily on their capacity to uncover manifolds in high-dimensional data spaces and project them into low-dimensional representation spaces. In this study, we investigate the structure and character of the manifolds generated by classical variational autoencoder (VAE) approaches and deep kernel learning (DKL). In the former case, the structure of the latent space is determined by the properties of the input data alone, while in the latter, the latent manifold forms as a result of an active learning process that balances the data distribution and target functionalities. We show that DKL with active learning can produce a more compact and smooth latent space which is more conducive to optimization compared to previously reported methods, such as the VAE. We demonstrate this behavior using a simple cards data set and extend it to the optimization of domain-generated trajectories in physical systems. Our findings suggest that latent manifolds constructed through active learning have a more beneficial structure for optimization problems, especially in feature-rich target-poor scenarios that are common in domain sciences, such as materials synthesis, energy storage, and molecular discovery. The jupyter notebooks that encapsulate the complete analysis accompany the article.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge