Mahnoosh Kholghi
Clinical Document Classification Using Labeled and Unlabeled Data Across Hospitals
Dec 04, 2018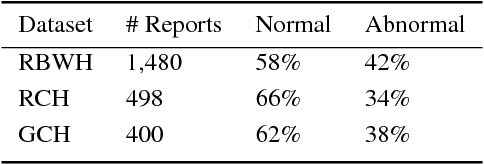
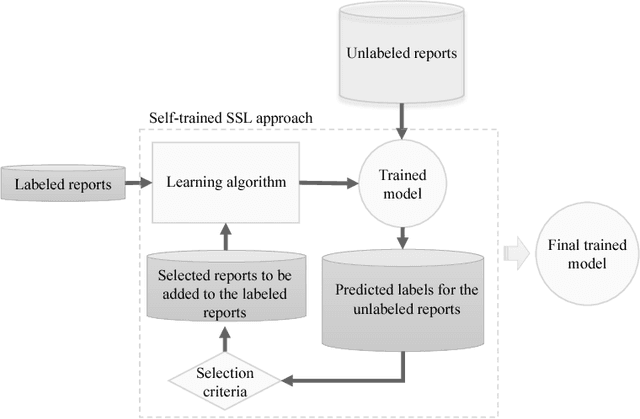

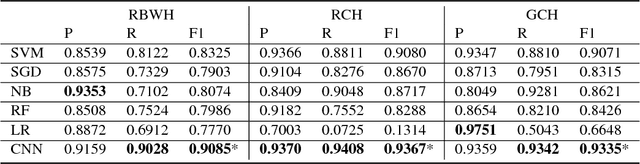
Abstract:Reviewing radiology reports in emergency departments is an essential but laborious task. Timely follow-up of patients with abnormal cases in their radiology reports may dramatically affect the patient's outcome, especially if they have been discharged with a different initial diagnosis. Machine learning approaches have been devised to expedite the process and detect the cases that demand instant follow up. However, these approaches require a large amount of labeled data to train reliable predictive models. Preparing such a large dataset, which needs to be manually annotated by health professionals, is costly and time-consuming. This paper investigates a semi-supervised learning framework for radiology report classification across three hospitals. The main goal is to leverage clinical unlabeled data in order to augment the learning process where limited labeled data is available. To further improve the classification performance, we also integrate a transfer learning technique into the semi-supervised learning pipeline . Our experimental findings show that (1) convolutional neural networks (CNNs), while being independent of any problem-specific feature engineering, achieve significantly higher effectiveness compared to conventional supervised learning approaches, (2) leveraging unlabeled data in training a CNN-based classifier reduces the dependency on labeled data by more than 50% to reach the same performance of a fully supervised CNN, and (3) transferring the knowledge gained from available labeled data in an external source hospital significantly improves the performance of a semi-supervised CNN model over their fully supervised counterparts in a target hospital.
The Benefits of Word Embeddings Features for Active Learning in Clinical Information Extraction
Nov 15, 2016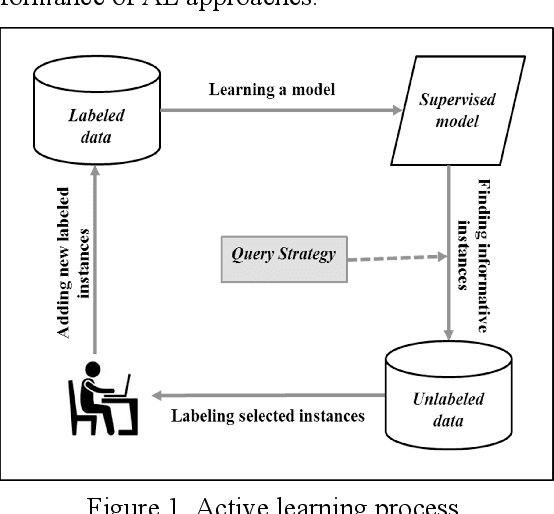
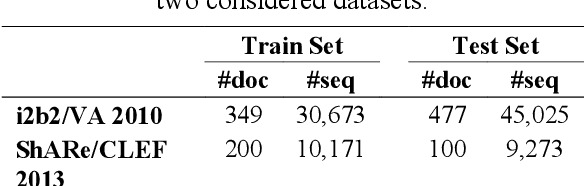
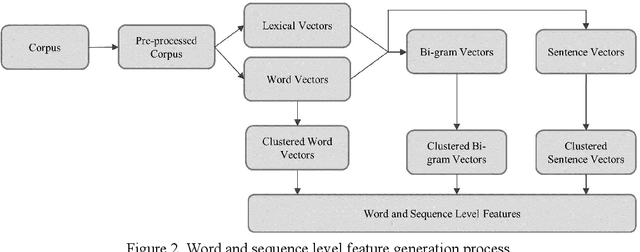
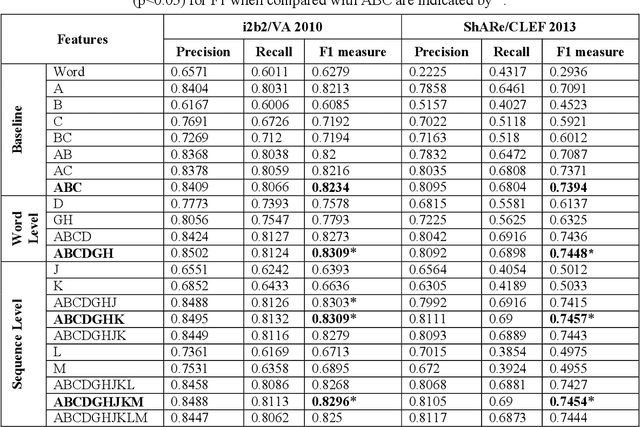
Abstract:This study investigates the use of unsupervised word embeddings and sequence features for sample representation in an active learning framework built to extract clinical concepts from clinical free text. The objective is to further reduce the manual annotation effort while achieving higher effectiveness compared to a set of baseline features. Unsupervised features are derived from skip-gram word embeddings and a sequence representation approach. The comparative performance of unsupervised features and baseline hand-crafted features in an active learning framework are investigated using a wide range of selection criteria including least confidence, information diversity, information density and diversity, and domain knowledge informativeness. Two clinical datasets are used for evaluation: the i2b2/VA 2010 NLP challenge and the ShARe/CLEF 2013 eHealth Evaluation Lab. Our results demonstrate significant improvements in terms of effectiveness as well as annotation effort savings across both datasets. Using unsupervised features along with baseline features for sample representation lead to further savings of up to 9% and 10% of the token and concept annotation rates, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge